Figures & data
Figure 1. HDAC activity in CLL B-cells, peripheral blood normal B-cells and umbilical cord blood B-cells. Mean HDAC activity for CD19+ purified CLL B-cells (n = 114), CD19+ purified peripheral blood normal B-cells (n = 15) and CD19+ purified umbilical cord blood B-cells (n = 15) is displayed with standard error of the mean (SEM) overlaid with scatter plot. Statistical differences were assessed using the Mann-Whitney non-parametric test.
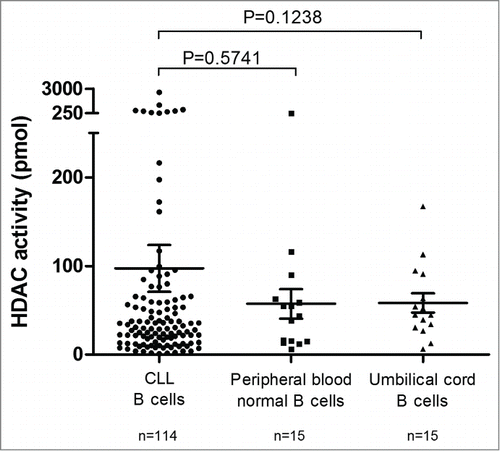
Figure 2. Prognostic power of HDAC activity. Deacetylation power was measured by enzymatic assay and cut-offs were optimized to maximize concordance with overall survival. Statistical differences between curves were calculated using the log-rank test. Statistical details can be found in Table S1. Kaplan-Meier curves showed the TFS (A) and OS (B) prognostic power of global HDAC activity. Cox regression was used to calculate hazard ratio for classical prognostic factors and HDAC activity in univariate (C) and multivariate analyses (D) for OS.
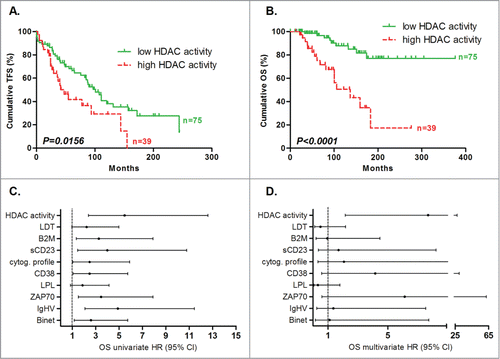
Figure 3. Refinement of prognosis sub-groups by HDAC activity for OS prediction. Patients classified in good prognostic subgroups by classical prognostic factors, including (A) ZAP70 – (n = 66), (B) LPL – (n = 67), (C) CD38 – (n = 62), (D) Binet stage A (n = 77), (E) IgHV mutated (n = 64), (F) B2M – (n = 54), (G) favorable cytogenetic abnormalities (n = 59), (H) lymphocyte doubling time >1 year (n = 73), and (I) soluble CD23 – (n = 61), were divided following their HDAC activity. The same was done for patients classified in poor prognosis subgroups by classical prognostic factors, including (J) ZAP-70 + (n = 48), (K) LPL + (n = 47), (L) CD38 + (n = 52), (M) Binet stage B - C (n = 37), (N) IgHV unmutated (n = 40), (O) B2M + (n = 22), (P) unfavorable cytogenetic abnormalities(n = 33), (Q) lymphocyte doubling time <1 year (n = 30), and (R) soluble CD23 + (n = 48). Statistical differences between curves were calculated using the log-rank test.
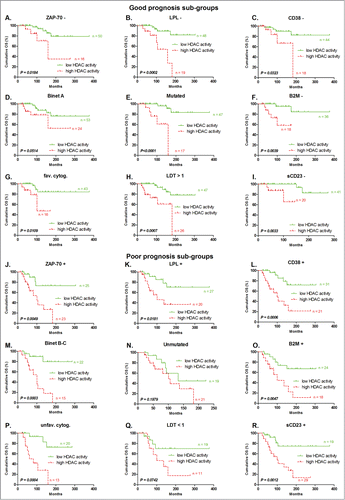
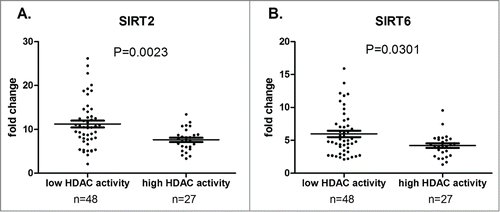
Figure 4. Correlation between HDAC RNA levels and global HDAC enzymatic activity. Mean HDAC mRNA levels (expressed as fold change) is displayed with standard error of the mean (SEM) overlaid with scatter plot for SIRT2 (A) and SIRT6 (B) in low (n = 48) and high (n = 27) HDAC activity groups. Statistical differences were assessed using the Mann-Whitney non-parametric test.