Figures & data
Figure 1. Schematic view to identify imprinted and monoallelically expressed genes. The pipeline shows the steps involved in analyzing and detecting imprinted and monoallelically expressed genes using RNA-Seq data and R scripts. Arrows show the data flow. Data sources (including RNA-Seq, SNP array data and external sources) are labeled by light gray boxes, while dark gray box shows the final result about imprinted and monoallelically expressed genes. UCSC: University of California Santa Cruz database.Citation17
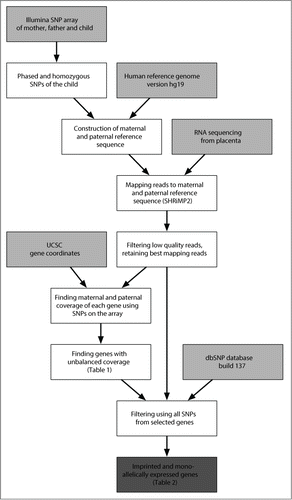
Table 1. Candidate imprinted genes after RNA-Seq and exome genotyping
Table 2. Imprinted and monoallelically expressed genes found in the current study
Table 3. Summary of detected imprinted and monoallelically expressed genes
Figure 2. Allele-specific expression in different placental samples. Each grid shows the allele-specific expression for each of the 12 imprinted and monoallelically expressed genes in different placental samples. The X-axis is the genomic position of the SNP and the Y-axis shows the percentage of maternal reads, with 100% refers to complete maternal expression, 0% corresponds to complete paternal expression and 50% refers to perfect biallelic expression. Statistically significant results are shown in red (maternal expression) and blue (paternal expression). From all non-significant results (depicted in black), only those are shown where the total number of RNA-Seq reads was at least 10. P001 - P014 are family trios analyzed.
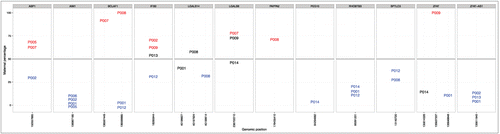
Figure 3. Density curve for gene expression. Red lines show the distribution of expression levels for all 12 imprinted and monoallelically expressed genes detected in the current study (see for details). Median expression of all genes is normalized to 1 (blue line).
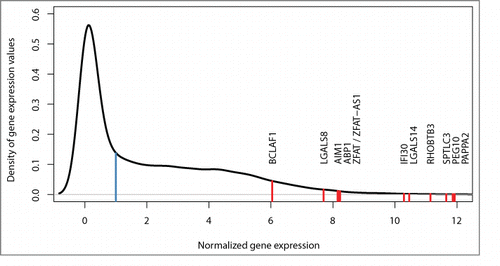
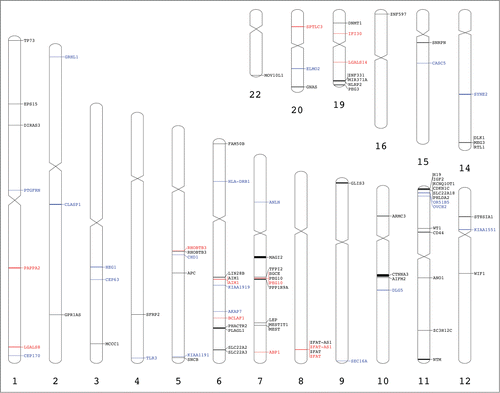
Figure 4. Genome visualization of the imprinted and monoallelically expressed genes. Previously reported placental imprinted genes are in black (Table S1), candidate imprinted genes are marked in blue () and imprinted and monoallelically expressed genes found in our analysis are marked in red (). This image was generated using Idiographica web service.Citation49