Figures & data
Table 1. Characteristics and Pb biomarker levels of study population.
Table 2. Percent of methylated cells at four candidate regions. Site-specific methylation and average of 4 CpG sites for repetitive elements, LINE-1, 4 sites within H19 and HSD11B2, and 3 sites for IGF2 are reported. Standardized values were adjusted for batch. LINE-1 was adjusted for batch by subtracting the 0% control value for each CpG site from the experimental plate each sample was run on. IGF2, H19, and HSD11B2 values were adjusted to a standard curve of control DNA (ranging between 0 and 100%) run for each CpG site on the corresponding experimental plate for each sample.
Figure 1. Simulation study findings. Bias in estimated coefficients and power to detect significant toxicant-DNA methylation associations from 5 approaches (see ), when data are generated to follow a similar structure to IGF (A and B) and HSD11B2 (C and D) methylation in the ELEMENT study (see Table S3 for simulation settings). For the batch-adjustment method using control samples, the scenario with standard curves having R2 = 0.99 (as we had in the ELEMENT study) is shown here. Bias and power are calculated from the average of 1000 data sets, each with 220 subjects.
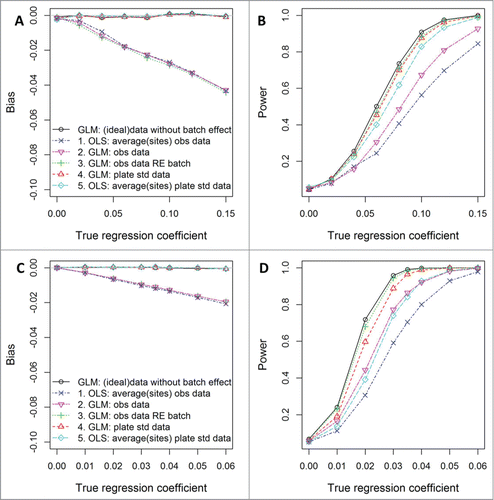
Table 3. Statistical models tested. Models used to examine associations between Pb biomarker levels and DNA methylation at LINE-1, IGF2, H19, or HSD11B2. Model #4 was considered the best approach though additional modeling types that are sometimes used with candidate gene methylation data were run for comparison purposes.
Table 4. IGF2 and HSD11B2 models. Estimates and p-values for the regression of methylation at IGF2 or HSD11B2 on each Pb biomarker are noted for a series of models described in . All models adjusted for sex, gestational age, and maternal folate intake, and models 2–4 also adjust for CpG site. Models included all subjects with available data (n = 223, 237, 221 for IGF2 models with umbilical cord blood, tibia, or patella Pb, respectively; n = 227, 241, 224 for HSD11B2 models with umbilical cord blood, tibia, or patella Pb, respectively).
Figure 2. Umbilical cord blood Pb and IGF2 methylation stratified by sex. Percent methylation in umbilical cord blood leukocyte DNA of the imprinted gene, IGF2, decreased with cord blood Pb solely in girls. After adjusting for gestational age and maternal folate intake, one natural-log unit increase in cord blood Pb was associated with a 3% decrease in IGF2 methylation among girls (P = 0.04) and the decrease among boys was not significant (P = 0.66). IGF2 data were first adjusted by a standard curve of known methylated or unmethylated controls from each experimental batch.
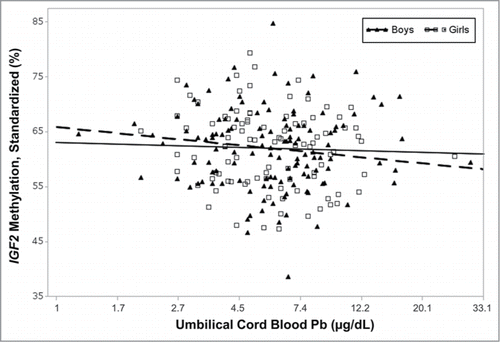
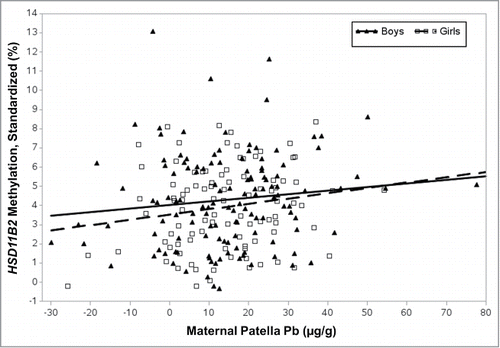
Figure 3. Maternal patella Pb and HSD11B2 methylation stratified by sex. Percent methylation upstream of the HSD11B2 promoter in blood leukocyte DNA increased with maternal patella Pb levels, indicative of in utero exposure. In a repeated measures model adjusting for gestational age and maternal folate intake, a 10 μg/g increase in maternal patella Pb of the girls was associated with a 0.23% increase in HSD11B2 methylation (near significant, P = 0.07). The increase was less notable among boys (P = 0.41). All methylation values were first standardized to known control curves from the same experimental batch.