Figures & data
Figure 1. Gene structure and protein interactions in yeast. (A) Phylogenetic analysis of putative MPC proteins in Arabidopsis. The amino acid sequences of these proteins were aligned by CLUSTALX, and the phylogenetic tree was constructed using the neighbor-joining method with the following parameters: bootstrap (replicates 1000), poisson model and complete deletion. The numbers at the nodes indicate the bootstrap values. (B) The interaction between the different MPC protein pairs. 32: pDEST32 for generation of the bait plasmid, 22: pDEST22 for construction of the prey plasmid. wt (pEXP22-RalGDS-wt), m1(pEXP22-RalGDS-m1) or m2 (pEXP22-RalGDS-m2) is control plasmid which shows strong, weak and undetectable interaction with pEXP32-Krev1, respectively. The transformed yeast cells grow on SC/-Leu-Trp (SC-L-T), SC/-Leu-Trp-Ura (SC-L-T-U), or SC/-Leu-Trp-His (SC-L-T-H) medium containing 50 mM 3-AT and aX-gal assay for β-galactosidase activity of the transformants grown on YPAD. (C) Test of protein interactions using a “swapped” 2-hybrid approach. The system is the same as shown in (B).
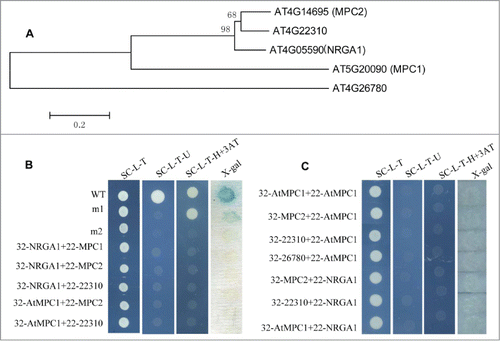
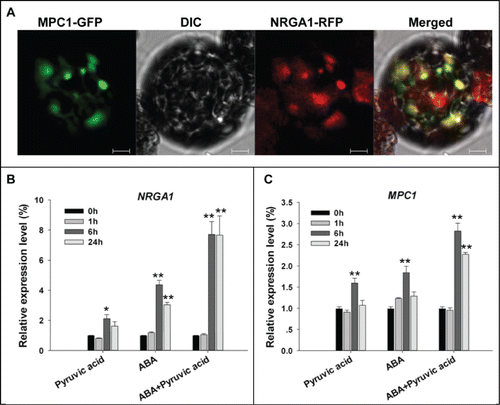
Figure 2. Subcellular localization of MPC1 and its gene transcription behavior. (A) Colocalization of GFP-tagged MPC1 and RFP-tagged NRGA1. The yellow signal indicates areas of overlap between the green and red signals. Quantitative RT-PCR of NRGA1 (B) and MPC1 (C) transcript under 200 μM pyruvic acid, 100 μM ABA or both were analyzed. Transcript levels relative to that of ACTIN2 are presented for each treatment. Each value represents the mean±SE of 3 independent measurements. The *, ** represent significant difference determined by the Student's t-test at P < 0.05 and P < 0.01 respectively. Bars = 10 μm.