Figures & data
Figure 1. Quantitative RT-PCR gene expression analyses of EXPANSIN A5 (AtEXPA5, At3 g29030). (A) HisAtRALF1-treated 10-d-old wild-type plants. Total RNA was extracted from roots after 30 min of treatment with 0, 0.1, 0.5 and 1 μM of the peptide. (B) Dark-grown 4-d-old wild-type plants treated with HisAtRALF1. Total RNA was extracted from hypocotyls after 30 min of treatment with 0, 0.1, 0.5 and 1 μM of the peptide. (C) Arabidopsis plants (10-d-old) treated with 100 nM of brassinolide (BL) for 6 h or pre-treated with 100 nM of BL for 5.5 h and treated with 1 μM of HisAtRALF1 for 30 min. Control plants were treated with H2O. Total RNA was extracted from the roots after each treatment. (D) AtRALF1-overexpressing (35S:AtRALF1), AtRALF1-silencing (irAtRALF1) and wild-type (WT) plants. Total RNA was extracted from roots of 10-d-old plants. Error bars indicate SD (* = p value < 0.05,** = p value < 0.01, t-test). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH, At1 g13440) expression was used as a control. All experiments were repeated at least 3 times (independent biological replicates).
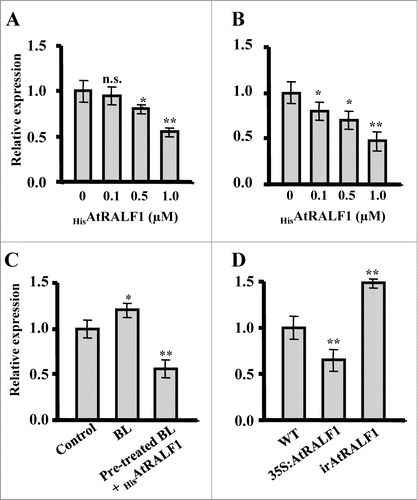
Figure 2. Schematic diagram of the AtRALF1 and BL gene regulatory network for cell expansion. Arrowed solid lines represent induction, and solid lines with bars at the end represent repression. Dashed lines represent the final effects of the actions of the genes in the cell expansion process. Dashed lines ending with arrowheads represent activation, and dashed lines ending with bars represent inhibition.