Figures & data
Figure 1. Schematic representation of Snail1, Snail2 and Twist1 domains (A) and the ubiquitin ligases involved in their degradation (B). (A) The position of the phospho-serine (PS) rich region, the nuclear export sequence (NES), the zinc finger domains (ZnF), the SLUG domain are shown for Snail1 and 2. The basic helix-loop-helix (bHLH) domain and the WR (tryptophan and arginine) motif is indicated for Twist1. (B) Fbxl5, Fbxl14 and Fbxw1/β-TrCP1 are represented. FBXL, F-box and Leucine-rich repeat Protein. FBXW, F-box and WD40 repeat protein. L, Leucine-rich repeat. W, WD40 repeat.
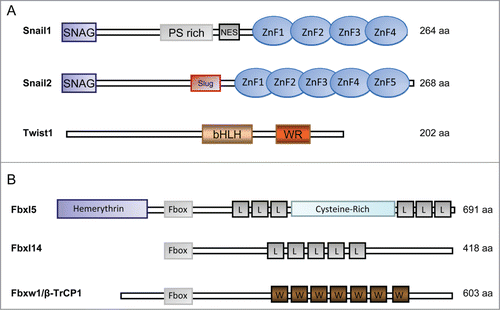
Figure 2. Composition of the E3 complexes targeting Snail1 and Snail2. The Skp-1-Cullin-1-F-Box (SCF) E3s, SCF-Fbxl14 and SCF-Fbxl5 are multimeric E3 ligases that mediates the ubiquitin (Ub) transfer from the E2 conjugating enzyme to Snail. SCF E3s are composed of the scaffold protein Cullin1 (Cul1), which interacts with Skp1 and the RING-finger protein Rbx1. The substrate binding affinity is mediated by the F-box protein (β-TrCP1, Fbxl14 and Fbxl5). The SCF-β-TrCP1 binds Snail1 only when double-phosphorylated by GSK-3β. Mdm2 is a single-subunit RING-finger E3 that targets Snail2 when p53 is bound.
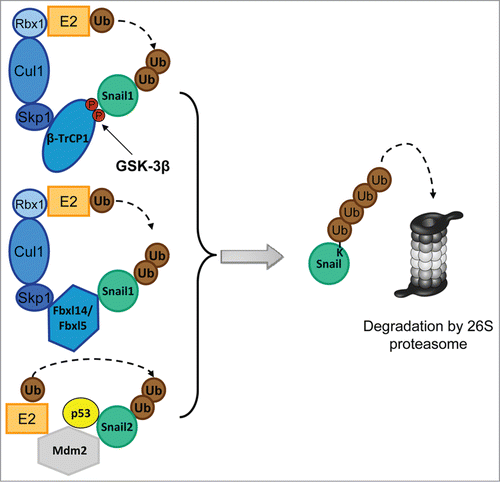
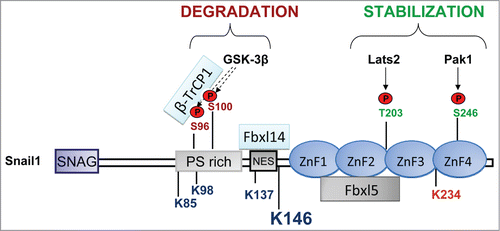
Figure 3. Snail1 phosphorylation sites linked to stabilization or degradation. Stabilizing phosphorylation sites are mainly localized in the C-terminus except the stabilizing ATM phosphorylation site in S100 and ERK2 phosphorylation sites in S82 and S104 (not indicated). The corresponding kinases are indicated by arrows. Phosphorylation sites promoting degradation are in the N-terminus (GSK-3β). The interaction with the F-box proteins Fbxl14 and β-TrCP1 is located in the N-terminus and with Fbxl5 in the C-terminus. The Lysines (K) that are ubiquitinated are indicated: K85, K146 and K234, were found to be specifically ubiquitinated by SCF-Fbxl5 and K98, K137 and K146 were modified by SCF-Fbxl14 or SCF-β-TrCP1. Although other K can be also modified, K146 is the best substrate of the 3 ligases and is the major ubiquitination site. K234 is marked in red because its modification by SCF-FBXL5 has a non-degradative role by decreasing DNA-affinity.