Figures & data
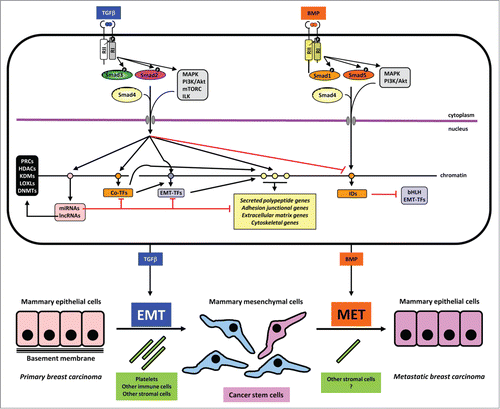
Figure 1. Mechanisms in TGFβ-induced EMT and BMP-induced MET. A mammary epithelial cell is shown on top with the 2 sister signaling pathways of TGFβ and BMP. These 2 growth factors are color coded to match the process of EMT and MET respectively. Signaling by the dimeric extracellular proteins is mediated by plasma membrane receptors (type II (RII) and type I (RI), which phosphorylate Smad proteins or activate other non-Smad signaling proteins (mainly protein kinases shown in gray boxes). Phosphorylation events on the type I receptor or on the Smads are shown with a black circled white P. Smads form complexes with Smad4 and together with non-Smad kinases transmit signals into the nucleus where different target genes are regulated positively (black arrows) or negatively (red arrows). Target genes of the pathways are symbolized as circles on the chromatin thread and are divided for simplicity and based on the text in 4 groups: a) miRNA and lncRNA genes (pink); b) co-transcription factor genes (orange), e.g., HMGA2, C/EBP or IDs in the case of BMP signaling; c) EMT-TF genes (purple); d) effector genes including secreted polypeptides, junctional, extracellular matrix and cytoskeletal components (yellow). MiRNAs are shown to negatively regulate various other target mRNAs. LncRNAs are shown to participate with chromatin regulators (black box on the left). These chromatin regulators participate in the chromatin-based epigenetic control of every step in the signaling and gene regulatory cascade (not shown). Make note of the sequential regulatory cascades whereby for example, the effector (yellow) gene is regulated by incoming Smad and non-Smad signaling, co-TFs and EMT-TFs, together with the conserted action of the lncRNA/chromatin factors (not shown). Such signaling mechanisms lead to EMT or MET leading to changes of cell architecture and differentiation as depicted at the bottom of the figure. Platelets and other cell types assist to the process of EMT, whereas the contribution of accessory cell types that promote MET is not well investigated (question mark). The generation of cancer stem cells is indicated with dark pink color and the metastatic breast carcinoma epithelial cells are colored as dark pink to indicate their development via MET from the CSCs. The latter is a speculative hypothesis and not yet demonstrated experimentally. Note the lack of normal basement membrane in the metastatic epithelial colony, which is also hypothetical and aims at emphasizing that the metastatic carcinoma cells and the epithelial cells in the primary cancer may not generate identically organized tissue architecture.