Figures & data
Table 1. Genomic location and orientation of PdL insertions in the mutant lines. Line ID - laboratory identifier of the mutant line. Designations in the second column are: XX:YYYYYYYY[+/-], where XX indicates the chromosome arm; YYYYYYYY indicates the Drosophila genome nucleotide number downstream of the PdL insertion; [+/-] indicates forward (5’-3’) or reverse (3’-5’) orientation of the PdL transposon in the chromosome. Genomic location and orientation are determined according to FlybaseCitation51 data released September 7th, 2010 (Dmel Release 5.30), sequenced strain y1; cn1 bw1 sp1 (Bloomington # 2057)
Table 2. Collection and FlyBase identification numbers (ID) of transgenic flies used in experiments. w* - unspecified allele
Figure 1. Locomotion parameters in flies with tissue-specific knockdown of the candidate genes (VDRC RNAi lines). GAL4/UAS-RNAi flies with spatially restricted knockdown (second column for each driver) derived by crossing elav/nrv2/appl/tsh-GAL4 drivers with VDRC RNAi lines. GAL4 controls without UAS-RNAi transgene (first, hatched columns) originate from cross of the same GAL4 drivers with host strain #60100 and are specific for each GAL4 driver. Also presented are locomotion parameters in UAS-RNAi controls descended from cross of CSBDSC with VDRC RNAi line for a gene indicated (CS, second columns) and in flies derived by crossing CSBDSC with host strain #60100 (CS, first, hatched column). Mean values with standard errors are shown. N = 40 for each data point. Significant difference from a corresponding control is indicated by filling (2-sided randomization test, 10,000 iterations, P < 0.05). Comparisons excluded from consideration (see text) are marked with asterisk. For details of genotypes see Methods and .
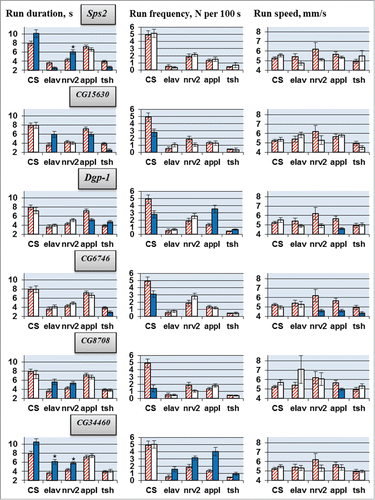
Figure 2. Locomotion parameters in flies with tissue-specific knockdown of the candidate genes (TRiP RNAi lines). Flies with spatially restricted knockdown derived by crossing elav/nrv2/appl/tsh-GAL4 drivers with TRiP RNAi lines. The host strain #36303 was used for all control crosses. Absence of a value indicates lethality or gross morphological effects of the knockdown. Other explanations, see legend for .
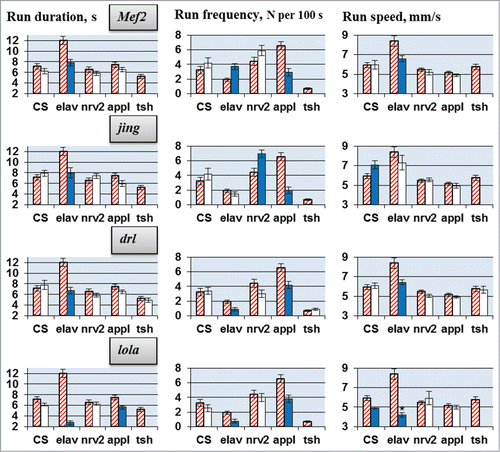
Figure 3. Courtship song parameters in flies with tissue-specific knockdown of the candidate genes. Flies with tissue-specific knockdown derived by crossing elav/nrv2/appl/tsh-GAL4 with VDRC/TRiP RNAi lines. N = 20 for each data point. Explanations are presented in legends for and .
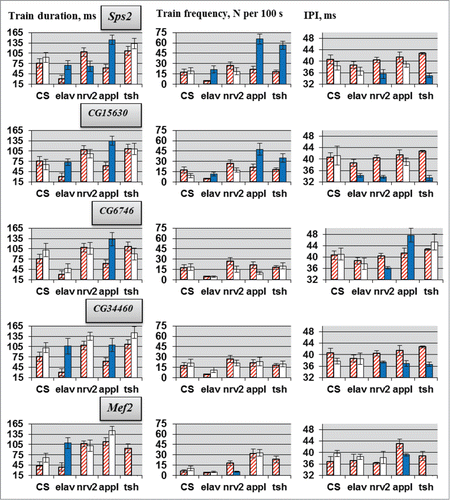
Figure 4. Expression level of Sps2, CG15630 and Mef2 genes in RNAi-mediated knockdowns relative to control. Mean expression ratios (knockdown/control) with standard errors are shown. Expression levels of all candidate genes are reduced in all experimental samples (2-sided randomization test, p < 0.05, REST 2009 software).
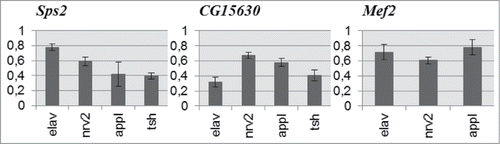
Figure 5. Motor parameters in flies with knockdown of CG15630 under the glia-specific repo-Gal4 driver. Glia-specific knockdown flies derived by crossing repo-GAL4 driver with VDRC RNAi lines #107797 for CG15630 and #105268 for Sps2. Control flies derived by crossing repo-GAL4 driver with host line #60100. Mean values and standard errors are shown. N = 40 for locomotor parameters, N = 20 for courtship song parameters. Significant difference from a corresponding control is indicated by filled columns (2-sided randomization test, 10,000 iterations, p < 0.05).
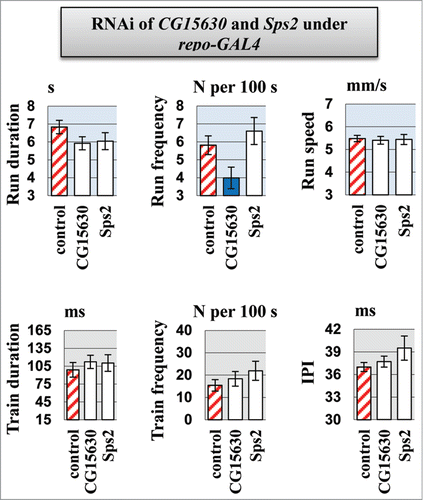
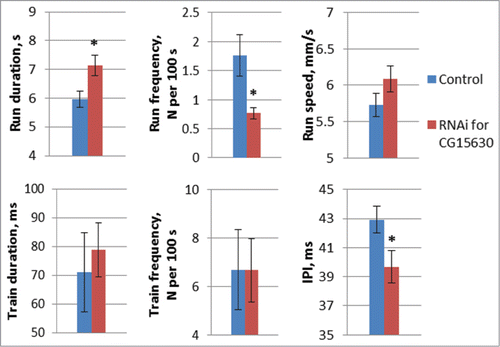
Figure 6. Motor parameters in flies with estrogen-induced knockdown of CG15630 in adults. Estrogen-induced knockdown flies derived by crossing GAL4.ER driver with VDRC RNAi line #107797 for CG15630. Control flies derived by crossing GAL4.ER driver with host line #60100. Both experimental and control flies were kept on estrogen-containing medium from the moment of imago eclosion. Mean values and standard errors are shown. N = 40 for locomotor parameters, N = 20 for courtship song parameters. Significant differences between RNAi and control data are marked with asterisks (2-sided randomization test, 10,000 iterations, P < 0.05).