Figures & data
Figure 1. (A) Clostridium difficile pathogenicity locus (PaLoc). Schematic organization of the C. difficile PaLoc, which is 19.6 kb in length. tcdA and tcdB (shaded in gray) are the 2 genes encoding the 2 large C. difficile toxins, TcdA and TcdB respectively. tcdR (shaded in black) encodes a positive regulator of transcription, whereas tcdC (shaded in black) encodes a putative negative regulator/modulatory protein. tcdE (shaded in white) encodes a holin protein. Adapted from J Dupuy et al.Citation8 J Med Microbiol 2008; 57: 685–90 and Carter et al.Citation5 J Bacteriol 2007; 189: 7290–7301. (B) Binary toxin locus (CdtLoc) from C. difficile 630. Schematic organization of the binary toxin locus (4.2 kb in length) from the binary toxin-negative C. difficile 630 strain. The CDT binary toxin-encoding pseudogenes, cdtAB, are shaded in gray. The response regulator gene, cdtR is shaded in black. The highly conserved 5′ and 3′ boundaries are also indicated. Adapted from Carter et al.Citation5 J Bacteriol 2007; 189: 7290–7301. (C) Binary toxin locus (CdtLoc) from C. difficile QCD-32g58. Schematic organization of the binary toxin locus (6.2 kb in length) from the binary toxin-positive C. difficile QCD-32g58 strain. The CDT binary toxin-encoding genes cdtA and cdtB, are shaded in gray. The response regulator gene, cdtR, is shaded in black. The highly conserved 5′ and 3′ boundaries are also indicated. Adapted from Carter et al.Citation5 J Bacteriol 2007; 189: 7290–7301. © American Society for Microbiology. Reproduced by permission of Becky Zwadyk. Permission to reuse must be obtained from the rightsholder.
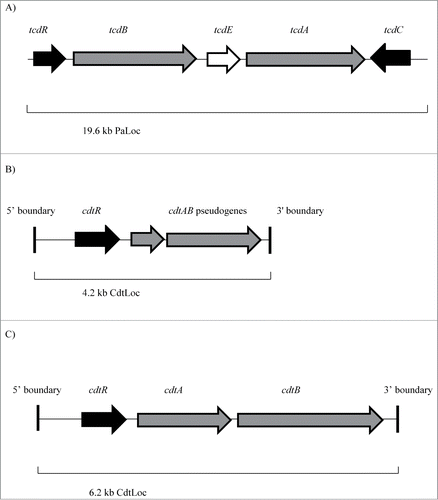
Figure 2. Narrow spectrum antimicrobial effects of thuricin CD. The effect of thuricin CD (90 μM) on family-level taxonomic distribution of the microbial communities present in model of the distal colon, expressed as percentage of total assignable sequences. Redrawn from Rea et al.Citation82 Proc Natl Acad Sci U S A 2011; 108: 4639–44. © Proceedings of the National Academy of Sciences of the United States of America. Reproduced by permission of Kay McLaughlin. Permission to reuse must be obtained from the rightsholder.
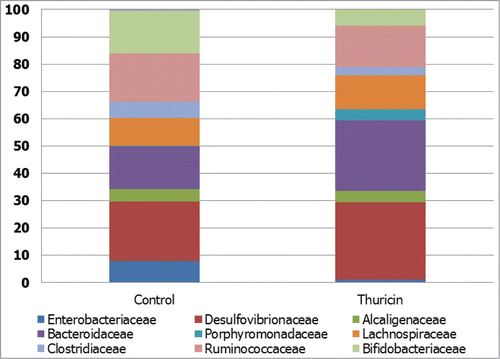
Figure 3. Broad spectrum antimicrobial effects of vancomycin, metronidazole and the bacteriocin lacticin 3147. The effects of the broad spectrum antimicrobials vancomycin, metronidazole and lacticin 3147 on phylum level diversity of gut communities in a model of the distal colon, expressed as percentage of total population of assignable tags. Other phyla: Actinobacteria, Spirochaetes, Lentisphaerae, and Tenericutes. Redrawn from Rea et al.Citation82 Proc Natl Acad Sci U S A 2011; 108: 4639–44. © Proceedings of the National Academy of Sciences of the United States of America. Reproduced by permission of Kay McLaughlin. Permission to reuse must be obtained from the rightsholder.
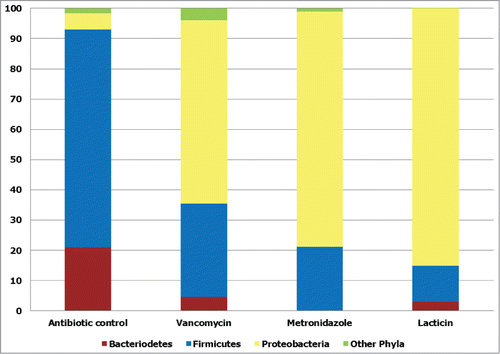
Table 1. Summary of minimum inhibitory concentration values (MIC) of anti-C. difficile antibiotics and bacteriocins. In vitro MICs of alternative antibiotics to metronidazole and vancomycin as well as as in vitro MICs of bacteriocins and bioengineered derivatives thereof against C. difficile strains as reported in the literature
