Figures & data
Figure 1. Late replicating chromatin on large chromosomes resides at the nuclear periphery. (A) Replication timing profile of a large (Chr4), intermediate (Chr14) and small (Chr21) chromosome in GM06990 lymphoblastioid cells (adapted from replicationdomain.org, visualized on http://genome.ucsc.edu; E = early, L = late replicating regions). Gaps mark centromere locations or unmapped NOR repeat regions. Above the tracks blue (=early) and red (=late) bars indicate the positions of BAC FISH probes used in the analysis. (B) FISH images illustrating the localization of an early (left 2 panels) and late (right 2 panels) replicating region. Each pair of images represents a single z-section through the nucleus showing one of the alleles. (C) Localization of early and late regions sampled from 6 chromosomes vis-à-vis the nuclear periphery. Early replicating loci exhibit lower peripheral association than late replicating loci (percentage of alleles contacting the lamina; Tukey boxplot). (D) Peripheral localization of early and late replicating loci as a measure of the size of the chromosome the probes are located on. Data is the same as in C. The exponential best fit curve illustrates the correlation between chromosome size and peripheral localization of late replicating loci.
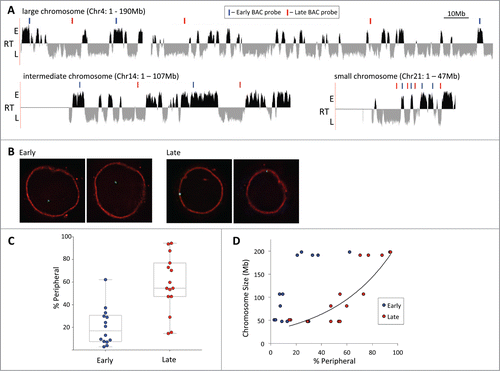
Figure 2. Genome association with the nuclear lamina in human lymphoblastoid cells by DamID. (A) Map of LaminB1 interactions with chromosome 3 (orange = enrichment, blue = depletion), above the replication timing profile (RT) of chromosome 3 (E = early, L = late). The LADs track in the center shows contiguous segments of prominent lamina association. DamID maps of chromosome 21 (B) and 22 (C) associations with LaminB1 above their respective replication timing profiles.
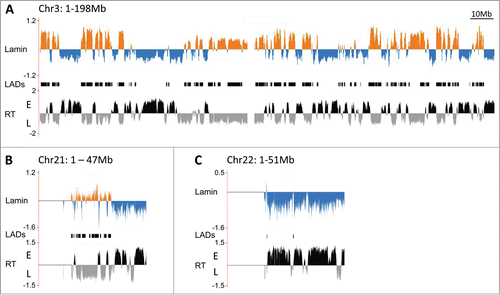
Figure 3. Late replicating loci associate with 3 repressive nuclear subcompartments. (A) The localization of several early and late replicating loci was determined simultaneously vis-à-vis the nuclear periphery (PH = peripheral heterochromatin), pericentromeric heterochromatin (PCH) and perinucleolar heterochromatin (PNH) by multicolor ImmunoFISH. Percent association of the loci to any of the 3 repressive subcompartments collectively (black) is significantly higher than association only with the PH compartment (blue). BAC probe identities: Chr17 A = RP11-142B17, B = RP11-42M14, Chr21 A = CTD-2059E17, B = RP11-315H16, C = CTD-2207L7, Chr22 = RP11-79G21. (B) Late loci in particular can localize to more than one repressive compartment at once. Venn diagrams show the association of two late and one early replicating region with the PH, PCH and PNH compartments. Early loci associate primarily with only one compartment. The size of the ellipses and their overlap is proportional to the percent association of a given locus with each compartment. BAC probe identities: Late Chr22 = RP11-79G21, Late Chr21 = RP11-315H16, Early Chr17 = RP11-142B17.
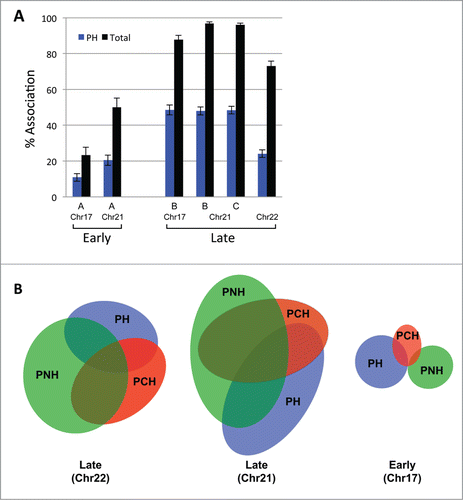
Figure 4. Repressive nuclear subcompartments exhibit redundancy. The positioning of a total of 4 different late replicating loci located on 3 different chromosomes vis-à-vis the PNH (A), PCH (B) and PH (C) subcompartments as well as their association to all 3 compartments collectively (D) (percent association) was assessed prior to and after 2 hours of actinomycin D treatment. Localization to repressive subcompartments overall is largely maintained, even after the disruption of nucleoli. Full color bars = prior to treatment; lighter shade bars = post actinomycin D treatment.
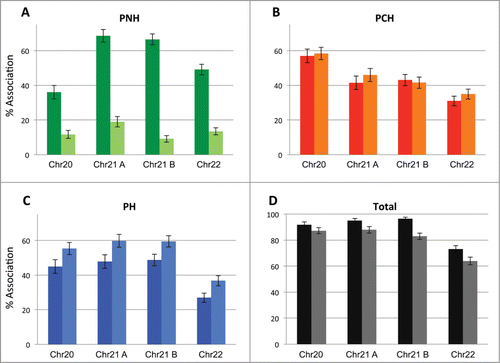
Figure 5. The late replicating genome localizes predominantly to 3 main repressive nuclear subcompartments: the nuclear periphery (PH, blue nuclear outline), pericentric heterochromatin (PCH, red) and perinucleolar heterochromatin (PNH, green). Our results suggest that a given late replicating region (yellow dots), particularly on small chromosomes, can associate with any (and multiple) of these subcompartments and relocalize from PNH to PH upon the loss of nucleoli (see arrow with check mark). While our results do not address the possibility of shuttling between the PCH and PH or PNH and PCH subcompartments (bidirectional arrows with question marks), work from other groups also suggest that localization between PH and PNH is interchangeable from one cell cycle to the next.