Figures & data
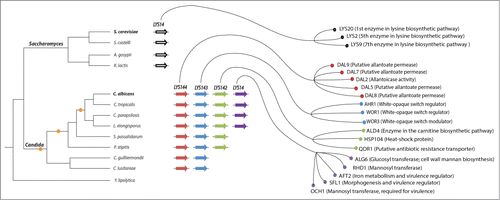
Figure 1. Transcriptional network expansion in the lineage leading to the human commensal and pathogenic yeast Candida albicans. The expansion and reshuffling of the Candida circuitry can be traced back to the successive duplications of a homolog of the S. cerevisiae transcription regulator LYS14. While LYS14 controls lysine biosynthesis in S. cerevisiae, each of the 4 resulting duplicated regulators in C. albicans has adopted a different role. A cladogram depicting the phylogenetic relationships among extant species of the Saccharomyces and Candida clades is shown to the left. The small orange circles in the cladogram represent the inferred gene duplication events that gave rise to the 4 C. albicans homologs. The thick arrows in the middle of the figure depict the genes encoding the LYS transcription regulators. Arrows of the same color represent orthologs based on phylogenetic reconstructions and synteny.Citation23 The right side of the figure shows a subset of each regulators’ target genes as determined by ChIP-chip.Citation23,27 Notice that the target genes included here are those that most likely contribute to the ability of C. albicans to colonize its mammalian host and cause disease.