Figures & data
Figure 1. STAT proteins interact with multiple chaperones to achieve nascent folding, activation, nuclear (and possibly mitochondrial) translocation, DNA binding, and possibly degradation.
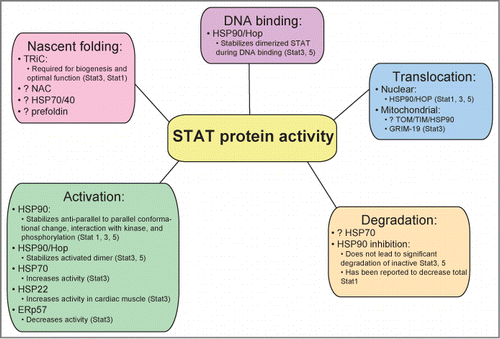
Figure 2. Interactions of chaperone proteins with Stat3 panel (A) and Jak2 panel (B). Stat3 requires interaction with TRiC for optimal biogenesis (portion 1). As most TRiC clients also interact with nascent polypeptide-associated complex (NAC), HSP70/HSP40, and prefoldin, we hypothesize that these chaperones are also involved in de novo Stat3 folding. Inactive Stat3 is found in large complexes in the cytoplasm containing chaperone proteins HSP90 and ERp57 (portion 2). Stat3 is recruited to activated cytokine receptors, and HSP90 is required for Stat3 interaction with JAK kinases, Stat3 phosphorylation, and dimerization of activated Stat3 (portion 3). It is thought that HSP90 stabilizes activated Stat3, making Stat3 phosphorylation and tail-to-tail dimerization more favorable. HSP90/Hop is required for Stat3 nuclear translocation (portion 4). HSP90/Hop co-localizes with Stat3, stabilizing Stat3 while it is bound to DNA (portion 5). Although data are limited, others have hypothesized that mtHSP90 is involved, in conjunction with TOM/TIM, with Stat3 mitochondrial translocation (portion 6). ERp57 activity within the ER decreases Stat3 activity in the cytoplasm (portion 7). The activity of other chaperone proteins, such as HSP70 and HSP22, is associated with increased Stat3 activity (not pictured). In panel B, tyrosine kinases are classic HSP90 clients that require interaction with HSP90 for de novo folding and maturation as well as activation. HSP70 plays an important role in the degradation of tyrosine kinases, which can occur at any stage of tyrosine kinase maturation or after activation. In the presence of HSP90 inhibitors tyrosine kinases are preferentially degraded.
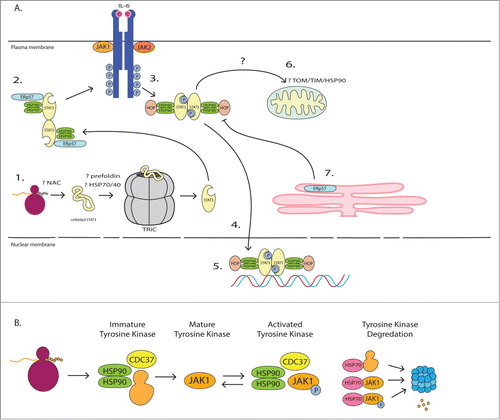
Figure 3. TRiC binds Stat1/Stat3 co-translationally and is required for Stat1/Stat3 synthesis in RRLs. In panel (A), TRiC was immunoprecipitated from rabbit reticulocyte lysate with a combination of antibodies to CCT2 and CCT5 (Anti-TRiC) or with a nonspecific control antibody (Human IgG) following translation of the indicated proteins in the presence ofCitation35S-methionine. Immunoprecipitates were separated by SDS-PAGE and autoradiographed. Half of each IP reaction prior to precipitation was run separately on SDS-PAGE and autoradiographed (Input). In panel (B), Stat1/Stat3 was translated in TRiC-depleted RRLs or following the addition of purified bovine TRiC in increasing amounts in the presence ofCitation35S-methionine followed by SDS-PAGE and autoradiography. The results shown are representative of 3 experiments.
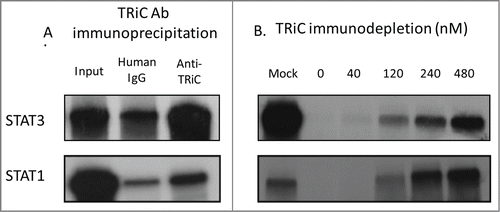
Table 1. Current and future strategies for modulating Stat3 activity for therapeutic benefit through targeting of chaperones.
