Figures & data
Figure 1. 10% SDS-PAGE analysis of monoclonal antibody h2E2 with or without treatment with peptide N-glycosidase-F (PNGase-F) to remove all N-linked glycans. After overnight treatment with PNGase-F, either 5 (lanes 2-4), 10 (lanes 5-7), or 20 μg (lanes 8-10) of h2E2 antibody were reduced and denatured in SDS and loaded onto a 10% acrylamide gel, followed by staining with Coomassie Blue R-250. Control, untreated h2E2 (“Con”) along with PNGase-F (“+”) or sham treated (“−”) monoclonal antibody are shown, with the dashed line added to aid visualization of the small differences in electrophoretic mobility observed after N-glycan removal by PNGase-F. The migration positions of the heavy chain, light chain, and PNGase-F enzyme are indicated on the right hand side.
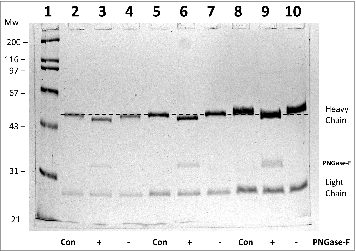
Figure 2. 10% SDS-PAGE gel analysis of TBP reduced and ABD-F labeled monoclonal antibody h2E2 heavy and light chains. 0.2 mg/ml antibody was first reduced with 4 mM TBP at 60°C for the times indicated in the figure, followed by alkylation with 4 mM ABD-F for 15 minutes at 22°C. Aliquots (5 μg) of the resultant samples were diluted in SDS-PAGE sample buffer and run on a 10% gel. Following photography under UV light to detect incorporated ABD-cys fluorescence (left hand side); the gel was then stained to measure total protein, and re-photographed (right hand side). Migratory positions of the heavy and light chains are indicated.
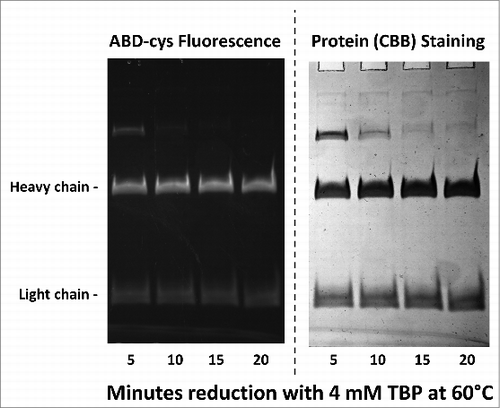
Figure 3. 5% acrylamide non-equilibrium pH gel electrophoresis (NEPHGE) analysis of monoclonal antibody h2E2. NEPHGE was performed as described in Methods, with the gel run for 90 minutes at 200 V. Aliquots of 5, 10 and 20 μg of h2E2 antibody were analyzed, and the region of the gel containing the separated variants of the monoclonal antibody is expanded on the right side of the figure. Note the separation of several groups of charged variants of the antibody by this technique. BioRad IEF gel standards are shown in the left lane of the gel.
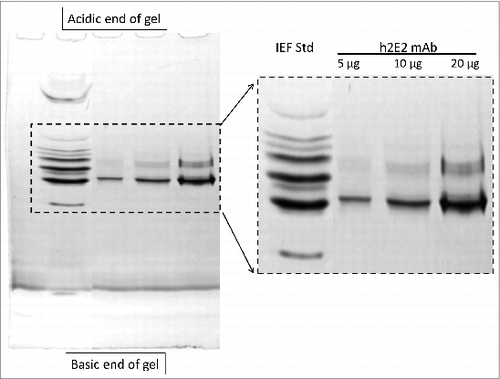
Figure 4. High performance strong cation exchange chromatography of the h2E2 antibody before and after treatment with carboxypeptidase B. 100 μg of h2E2 antibody was injected, and eluted with a gradient of NaCl in MES buffer. Note the disappearance of peaks 1 and 2 after removal of the C-terminal lysine residues. These peaks represent h2E2 antibody molecules containing 1 or 2 lysine residues on the C-termini of the 2 heavy chains in the antibody (which, after removal of lysine by carboxypeptidase elute with the main peak labeled ‘0” eluting at approximately 11 minutes).
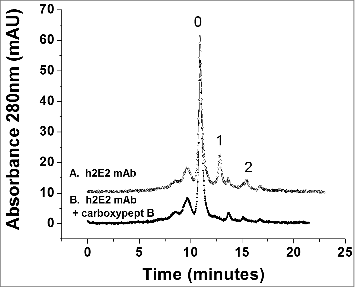
Figure 5. Reducing 10% SDS-PAGE analysis of h2E2 Fab, mAb and the Endo Lys-C digest. 5 μg of Fab, 6 μg of h2E2 mAb and 6 μg of Endo-Lys-C digested mAb were loaded onto lanes 2-4, respectively. Note the purity and lack of high molecular weight impurities in the Fab preparation. Migration positions of the reduced heavy and light chains, as well as the reduced Fab and Fc fragments, are indicated.
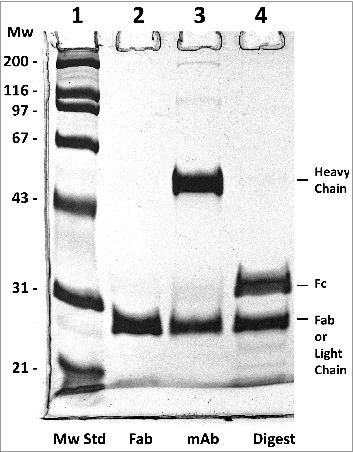
Figure 6. High performance strong cation exchange chromatography of the Fab fragment derived from the h2E2 antibody. 100 μg purified Fab fragment was injected, and eluted with a gradient of NaCl in MES buffer at 22°C. Note the relative lack of charge heterogeneity compared to the intact h2E2 antibody ( and the inset), indicating much of the charge heterogeneity resides in the Fc portion of the h2E2 antibody. For comparative purposes, the position and pattern of the intact antibody on the same column using the same buffers and gradient is shown in the inset (offset in the y-direction for clarity).
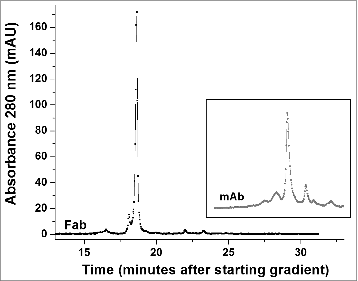
Figure 7. Intrinsic h2E2 antibody tryptophan and tyrosine fluorescence quenching by cocaine binding. Shown are emission spectra of 5 nM h2E2 antibody both before and after addition of 100 nM cocaine, with excitation at either 280 nm (tyrosine and tryptophan) or 295 nm (tryptophan). Note the decrease in emission (quenching) caused by cocaine, but little if any change in the emission maximum (near 330 nm in both cases).
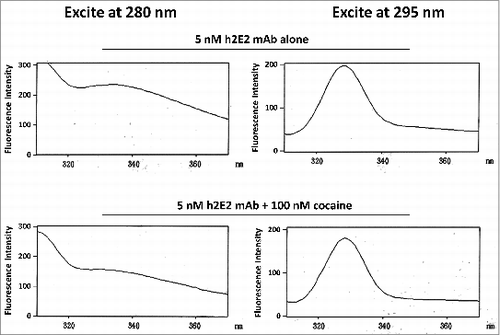
Figure 8. Titration of 5 nM h2E2 antibody with cocaine, with excitation at both 280 nm and 295 nm. Only data represented by filled symbols were used for fitting to obtain the indicated sigmoidal curves to derive the EC50 values. Note the y-axis brake in scale to allow visualization of the quality of the fitted curves for both sets of data on a single plot. KD values were calculated from the EC50 values and the antibody or Fab concentration as described in Methods. For the experiment shown in the figure (using the intact h2E2 mAb and 295 nm excitation), KD = EC50 – (0.5 × 2 sites/mAb × 5.0 nM mAb) = 9.4 nM – 5.0 nM = 4.4 nM = KD for cocaine binding to the h2E2 monoclonal antibody.
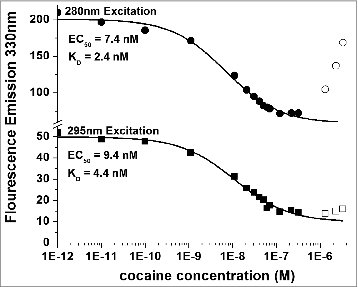
Table 1. Summary of cocaine and cocaine metabolites binding to both the intact antibody and the Fab fragment of the h2E2 mAb, as measured by quenching of intrinsic protein fluorescence.
