Figures & data
Figure 1. RNAi pathways in insects. The figure shows the biogenesis pathways for siRNAs, miRNAs and piRNAs in insects. In the siRNA pathway, the RNA transcript from a DNA construct encoding hairpin loop sequences, or from a virus-derived dsRNA will trigger siRNA generation in the cytoplasm, resulting from the recognition and dicing by Dicer 2. The siRNA duplexes will then load into the AGO 2 - RISC complex. The guide strand of siRNA is maintained in the RISC and will lead it to the target mRNA for degradation/cleavage or translation repression. For endogenous miRNAs, pri-miRNA is transcribed from genomic DNA, and processed by Drosha and cofactor Pasha (DGCR8) into the pre-miRNA. Next, the pre-miRNA will be transported into cytoplasm by exportin 5 and Ran-GTP, and cleaved by Dicer 1 into miRNA duplexes. Similar to siRNAs, the miRNA duplexes will incorporate into the AGO 1 - RISC complex, followed by target mRNA cleavage/degradation or translation repression. In the piRNA pathway, piRNA precursors are transcribed from piRNA clusters in the genomic DNA sequences. The antisense transcript could transport into cytoplasm, processed by series of proteins of the Piwi-group, in Zuc dependent or independent manner and incorporated into the PIWI-RISC complex, or into the AUB-RISC complex in the ping-pong cycle. The sense strand, which is targeted by the antisense strand in AUB-RISC, will then be cleaved and incorporated into AGO 3 to target the antisense strand; hence the ping-pong cycle.
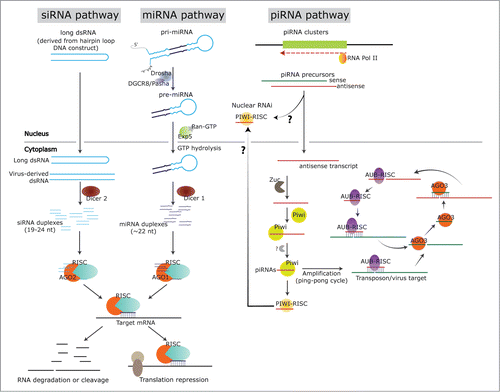
Figure 2. Mapping summary of vsRNA reads against HoCV-1 genomic RNA. The numbers of vsRNA reads mapped along the Y -axis and the HoCV-1 genomic RNA are represented across the X -axis. The gray colored peaks represent the positive-strand small RNAs while the red colored peaks represent the negative-strand small RNAs. This figure is adapted from Nandety et.,al 2013Citation23 with permission from Virology, Elsevier Limited. This is just one example, several publications describe the distribution of small RNA reads on different viruses.Citation22,24
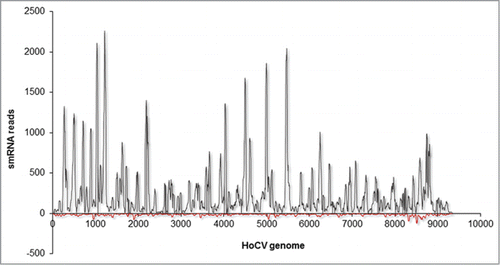
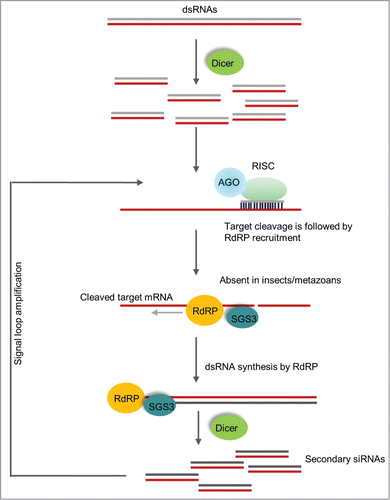
Figure 3. RNA dependent RNA polymerase amplification of siRNAs. The figure shows the signal amplification by RdRP and processing of siRNAs in plants. In this mechanism, Dicer processed primary siRNAs from an exogenous dsRNA guide the target mRNA cleavage. The cleaved mRNA will then be used as template for RdRP to produce dsRNA and targeted by Dicer, initiating the amplification cycle. This type of robust amplification is absent in hemipteran insects but present in plants and C. elegans.