Figures & data
Figure 1. Loss of CREB and CREM impairs the proliferation of striatal precursors. (A–B) Quantitative analysis of total cells in high (A) and clonal (B) density neurosphere cultures established from dissociated GE of E13.5 embryos with the indicated genotypes. Data represent means ± SEM in at least 3 independent experiments. Asterisks indicate significant differences versus control (Ctr; ANOVA) * P <0 .05; ** P<0 .01. (C) Representative images illustrating clone size and morphology.
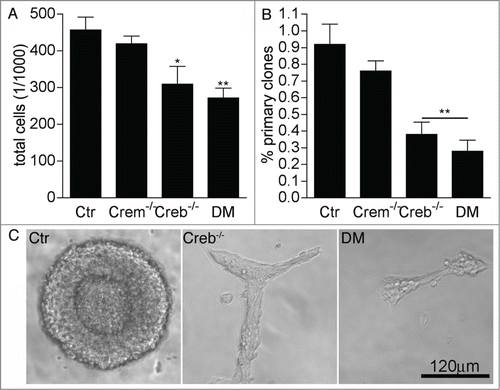
Figure 2. Effect of CREB and CREM on number and cycling rate of EGFRhigh precursors. (A) Representative FACS plots illustrating EGFRhigh cells in DIV1 control (Ctr) and DM cultures, pre-treated with EGF before staining with EGF-Alexa488, as negative control, or left untreated as indicated. (B–E) Quantitative analysis of EGFRhigh cells in the cultures established from the GE of E13.5 embryos with the indicated genotype measured after the indicated day in vitro (DIV). In (D) the culture medium was replaced 24 hours after plating. (F) Quantitative analysis of BrdU incorporation and expression of the given markers in EGFRhigh cells isolated from DIV2 cultures of Ctr and DM GE. Data represent means ± SEM in at least 3 independent experiments. Asterisks indicate significant differences vs. Ctr (Student's t-test) *P <0 .05; **P <0 .01. (G) Fold increase in cell number from DIV0 to DIV5 neurosphere cultures established from dissociated E13.5 GE cells of embryos with the indicated genotypes.
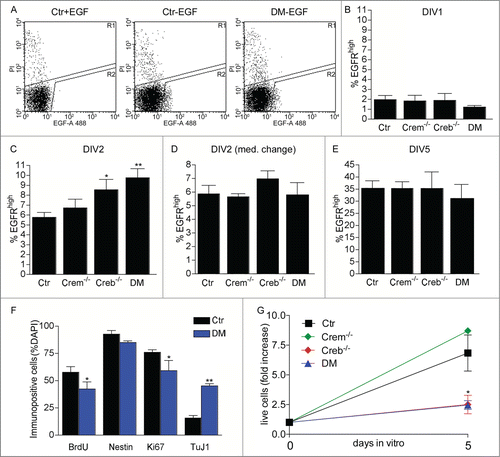
Figure 3. Effect of CREB and CREM on the ability of precursors isolated from the E13.5 GE to form primary and secondary clones. Analysis of clones obtained from EGFRhigh and/or EGFRlow cells sorted at DIV1 and DIV2 (A and D) and DIV 3-5 (E and H) from cultures of the GE from E13.5 embryos with the given genotypes. Graphs illustrate quantitative analyses of primary (A, B, E, F) and secondary (D and H) clones as well as clone size (C and G). Data represent means ± SEM in at least 3 independent experiments. Asterisks indicate significant differences versus Ctr (ANOVA) *P <0 .05; **P <0 .01.
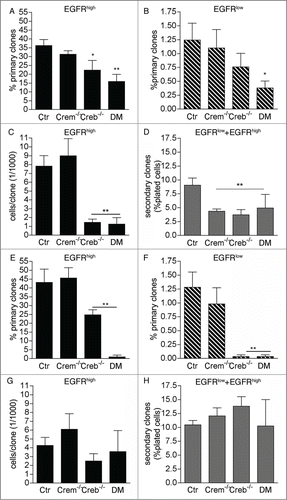
Figure 4. Analysis of phospho-CREB and PCNA expression in the coronal sections of the E13.5 telencephalon. Representative confocal micrographs illustrating the germinal regions of the GE and cortex from control (Ctr) and DM embryos upon double immunostaining with phospho-CREB (P-CREB, red), PCNA (green) and DAPI (blue) counterstaining of the nuclei. The apical side is shown on the upper side. Arrows and arrowheads indicate double and single immunopositive cells, respectively. Scale bar is 30 μM.
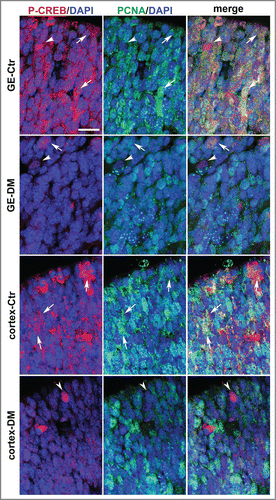
Figure 5. Ablation of Creb and Crem leads to reduced proliferation in the GE but not in the cortex of E13.5 embryos. (A–D) Representative micrographs of coronal telencephalic sections obtained from control (Ctr; A and B) and DM (C and D) embryos upon immunostaining with BrdU (A and C) and phospho-H3 (B and D) antibodies. (E) Quantitative analyses of the immunostaining with BrdU (Ea and Ec) and PH3 (Eb and Ec) antibodies in the GE (Ea and Eb) and in the cortex (Ec and Ed). Scale bar is 60 μm; N = 3–4. Asterisks indicate significant differences vs. control (Ctr) counterpart (Student's t-test) *P < 0 .05.
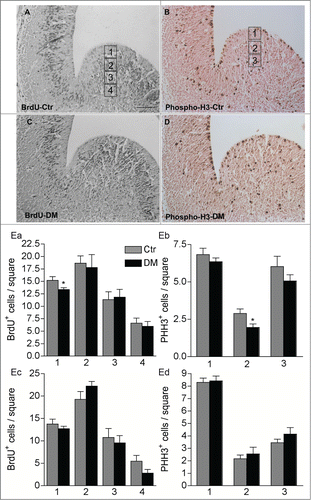
Figure 6. Ablation of CREB and CREM results in decreased histone acetylation in the E13.5 telencephalon. (A) Western blot analysis of GE (left column panels) and cortical protein extracts (right column panels) showing analysis of αlpha-tubulin, total histone H2B, acetylated histones H3 and H4. (B) Quantification of western blot analysis of the GE (Ba and Bb) and cortex (Bc and Bd). Bands were normalized against H2B (Ba and Bc) and α Tubulin (Bb and Bd). Note that ablation of CREB leads to a stronger alteration of histone acetylation in the GE (Ba and Bb) than in the cortex (Bc and Bd). Asterisks indicate significantly different from the respective control (Ctr) (ANOVA) * P < 0.05; **P < 0.01.
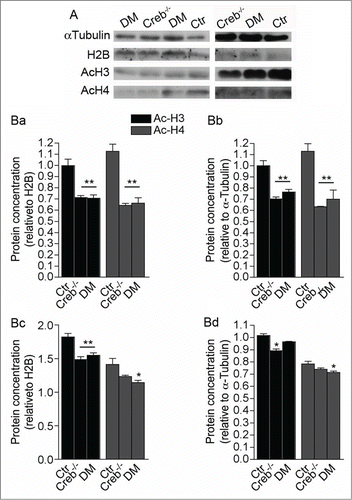
Figure 7. Blocking histone deacetylation rescues the ability of neural precursors to form clones. (A) Representative micrographs of coronal telencephalic sections obtained from control E13.5 embryos showing the effect of TSA on the levels and distribution of acetylated histone 3 (AcH3), acetylated histone 4 (AcH4) and histone 3 (H3) immunoreactivity in the GE and in the cortex. Pregnant females were injected with TSA at 11 d.p.c. and every 12h for 48h before analysis. V = Ventricle. Scale bar is 30 mM. (B) Quantitative analyses of the number of primary clones obtained from EGFRhigh and EGFRlow cells isolated at DIV 1 cultures established from E13.5 embryos with the indicated genotypes after TSA treatment.
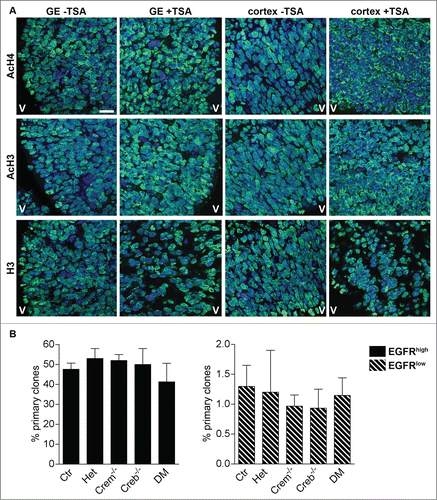
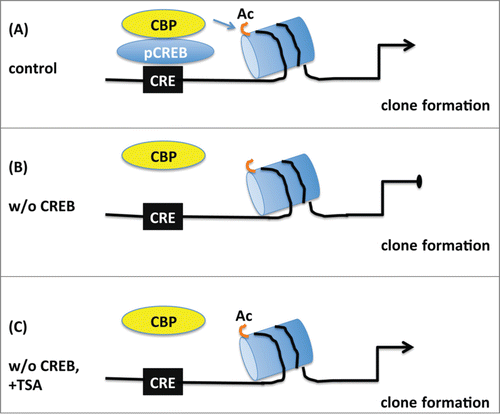
Figure 8. Schematic model depicting the proposed role of CREB in the proliferation of embryonic neural progenitors. (A) Under normal conditions phosphorylated CREB (pCREB) binds to cAMP-responsive element (CRE) and recruits CBP promoting histone acetylation (Ac) of targets genes which in turn lead to clone formation. (B) Upon CREB ablation, CBP is not capable of maintaining histone acetylation and clonogenic activity of neural progenitors is impaired. (C) Treatment with the histone deacetylase inhibitor TSA prevents the loss of histone acetylation and rescues the deficit in clone formation observed in the absence of CREB suggesting that CREB regulates proliferation essentially by modification of chromatin structure.