Figures & data
Figure 1. Individual RG cells were labeled using electroporation. (A) In embryonic animals, ex ovo electroporation was used. Subsequently, brains were either cultured as coronally sectioned slices or cultured in the whole embryo. Electrodes were applied following a dorso-ventral orientation to label cells in the dorsal cortex. (B) In adult animals, the brain was removed, hemisected and embedded in agarose. Electrode positioning following a medio-lateral orientation to label cells in the DVR and striatum. After electroporation, brains were cut into slices and organotypic slice cultures prepared.
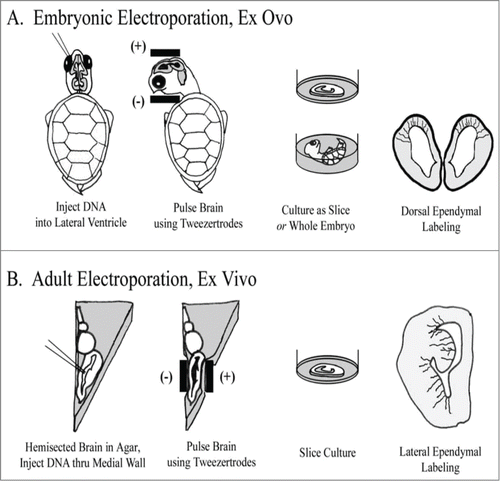
Figure 2. Electroporation of the embryonic turtle telencephalon with an EGFP-expressing plasmid labels many cells with RG morphology. (A) At 24 hours post-electroporation of a stage 20 embryo, an array of EGFP+ cells in the dorsal cortex show RG morphology. (B) At higher magnification a variety of morphologies are seen. RG cells in different phases of the cell cycle are apparent (B1, B2). Small expansions are often present along the length of the pial process (B3, B4). A smaller number of EGFP+ cells in the VZ do not appear to have a pial process. These may be newly generated neurons, intermediate progenitor cells, or RG cells. At later stages (stage 23), cells are found with bifurcated or branched pial processes, and ‘hairy’ or more lamellate fibers (B5). (C) A variety of ventricular contacting processes are seen. Some processes are typical for embryonic RG cells seen in rodents (C1, C2) while others are more complex (C3, C4). (D) RG cells exhibit different heterogeneous morphologies. D1, RG cells in younger animals often have a smooth pial process. In many cases expansions bud off of the process (D2) or in line with the process (D3). As development proceeds RG cell pial fibers exhibit more mature ‘lamellate’ fibers (D4). Scale bars: A, 50 μm; B, 10 μm; C,D, 10 μm.
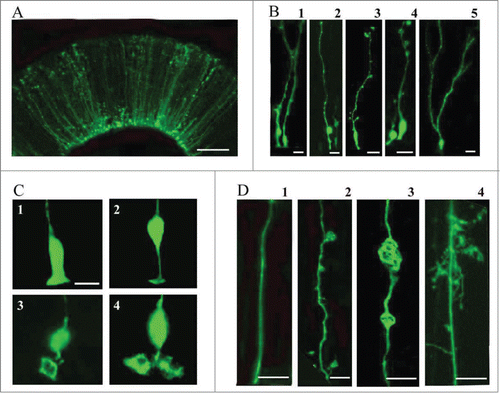
Figure 3. RG cells migrate, exhibit cellular movements, and proliferate in organotypic slice cultures and in whole embryo cultures. (A) RG cells in the embryonic turtle telencephalon labeled through electroporation. Shown are 2 time points, 12 hours and 36 hours. Electroporated cells were adjacent to the ventricle at early time points (A1), but at later time points, some RG cell bodies had translocated away from the ventricular surface (A2). (B and C) Slice cultures allowed to incubate for up to 4 weeks contained cells with mature neuronal morphologies. In C, the neuron (arrow) is located just superficial to the proliferating area of ependymal cells (arrowhead). (D and E) Representative examples of dorsal cortex that were incubated in whole embryos cultures for 12 hours (D) and 3 d (E). At 12 hours, RG cell bodies were close to the ventricle. At 72 hours, many electroporated cells had migrated or translocated into the cortex (arrows in E). (F) Whole embryos cultured with a pulse of BrdU. Left panel shows a single electroporated RG cell (green). The second panel is a higher power image of the same cell. BrdU staining revealed many mitotic cells (red), including GFP+ RG cells that were BrdU+ (green, arrowheads). The right panel is a negative control BrdU staining. DAPI stain (blue) labels all nuclei. Scale bars: A, B, C, 20 μm; D, E, 100 μm; F, 5 μm.
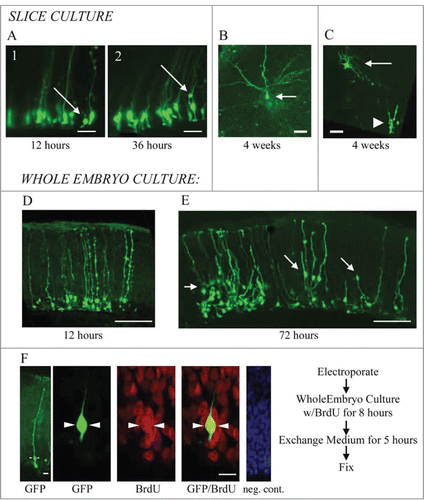
Figure 4. M-phase cells in the embryonic turtle VZ express the RG lineage marker phosphorylated vimentin (4A4) and reveal RG cell morphology. (A) En face cortical slab stained with syto-11 (green) and 4A4 (red). All M-phase cells at the ventricular surface express 4A4. (B) 4A4 robustly labels M-phase cells throughout mitosis. (C) In coronal sections 4A4 labeled mitotic cells lining the ventricle that possessed pial fibers coursing out through the parenchyma. Pial fibers were most robust in prophase, but a thin process remained throughout division. (D) A brief BrdU (blue) pulse shows that S-phase cells are found in an abventricular position at the top of the VZ while 4A4+ cells mitotic cells were located at the ventricular surface, indicating interkinetic nuclear migration in the developing turtle telencephalon. Scale bars: A, 20 μm; B, 5 μm; C, 10 μm; D, 20 μm.
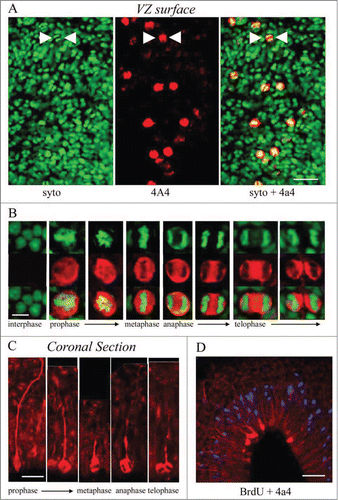
Figure 5. Tbr2-expressing intermediate progenitor cells are present in the developing turtle telencephalon. (A) A coronal section of a stage 20 turtle embryo immunostained with anti-PCNA (green) and anti-Tbr2 (red), and costained with DAPI (blue). (B and C) The distribution of Tbr2+ cells (red) differs in the dorsal cortex versus the dorsal DVR. Tbr2+ cells are distributed throughout the VZ in the dorsal cortex, but concentrate in a band superficial to the VZ in the DVR, as is seen in the developing mammalian neocortex.
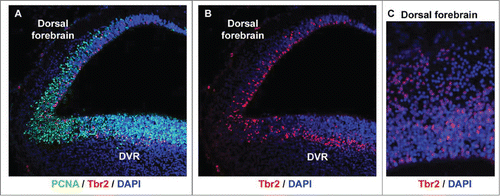
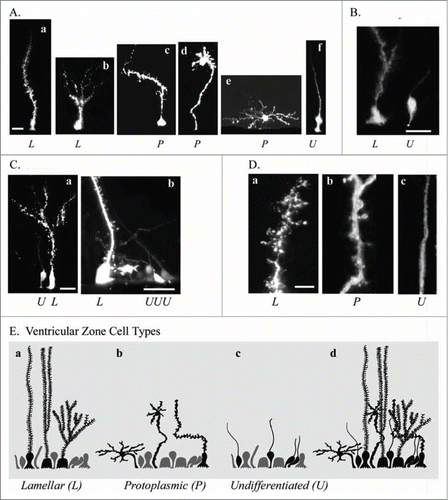
Figure 6. Electroporation of the adult turtle telencephalon reveals heterogeneous RG cells that we grouped into 3 categories distinguishable by their pial fiber morphology. Lamellate RG cells (L, Aa, Ab); Protoplasmic RG cells (P, Ac−Ae); and Undifferentiated RG cells (U, Af). (B) Further examples of the 3 cell types identified by letter under each image. (C) Lamellate RG cells have pial fibers possessing ‘hairy’ fine extensions, and a pial fiber that in some cases had multiple branches within the parenchyma. (D) Comparison of the pial fiber of the 3 cell types in higher magnification images. Protoplasmic fibers had many smooth expansions. Cell bodies were located at the ventricle and away from the ventricle. Protoplasmic RG cells had the most diverse cellular morphologies. Undifferentiated fiber types were smooth and traceable through the pyramidal cell layer and for several hundred micrometers into the parenchyma. They arose from smaller cell bodies most frequently found close to the ventricle (B and C). (E) Schematic showing the 3 classes of RG cells and their overlapping distribution (Ed). We hypothesize that undifferentiated RG cells retain the capacity for proliferation. Scale bars: A, B, C, 10 μm; D, 3 μm.