Figures & data
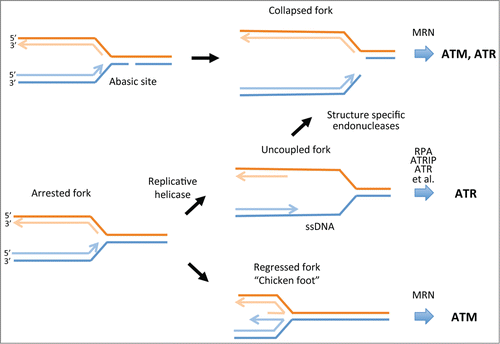
Figure 1. Fate of DNA replication forks in response to DNA damage or replication stress. Some forms of DNA damage such as abasic sites that may arise through the action of ROS can directly cause replication fork collapse. Such events trigger the activation of ATM or ATR, depending on the processing of the end by the MRE11/RAD50/NBS1 (MRN) complex or other DNA damage response proteins that may be recruited to the collapsed fork. Uncoupling of polymerase and helicase complexes at arrested forks can lead to the formation of stretches of ssDNA. These become coated with RPA, which then recruits ATRIP and ATR and a number of other proteins to activate the ATR-CHK1 protein kinase cascade. If ssDNA is insufficiently protected by RPA, structure-specific endonucleases such as MUS81 can cleave ssDNA to collapse the fork. Arrested forks may also undergo regression to produce the so-called chicken foot structure. This response potentially enables replication complexes to bypass DNA lesions or difficult to replicate regions. This structure also produces a substrate for the 3′–5′ exonuclease activity of MRE11 that can activate ATM. Regressed forks can also be resolved by a number of other pathways that generate DSBs or ssDNA to restart DNA synthesis. ATM, ataxia telangiectasia-mutated; ATR, ATM and Rad3 related; CHK1, checkpoint kinase 1; DSBs, DNA double-strand breaks; ROS, reactive oxygen species; RPA, replication protein A; ssDNA, single-stranded DNA.