Figures & data
Figure 1. LC3 immunohistochemical staining shows a punctate pattern in wild-type MEFs and a homogenous staining pattern in atg7−/− MEFs. (A) Western blot of ATG7, SQSTM1/p62, LC3-I and LC3-II and MAPK14/p38 in wild-type and atg7−/− MEFs. (B) Representative images of immunohistochemical detection of LC3 in wild-type and atg7−/− MEFs that were cultured in replete medium (untreated) or starved in EBSS for 2 h. Antigen retrieval was undertaken in Tris-EDTA, pH 9. Images were taken at 100x magnification and the scale bars in each panel represent 20 µm.
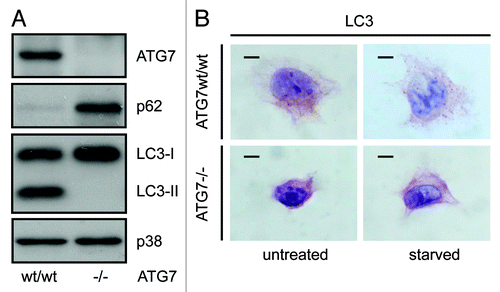
Figure 2. Detection of a punctate LC3-staining pattern in murine wild-type pancreas and uniform staining in atg7−/− tissue. Immunohistochemical staining for ATG7, LC3, SQSTM1/p62 and ubiquitin in murine wild-type pancreata (top two rows) and in consecutive sections from pancreata with mosaic deletion of ATG7 (atg7−/−, bottom two rows). Panels with straight borders are complete, i. e. not cropped and representative overview images taken at 40x magnification with a scale bar (straight) that represents 100 µm. Panels with dashed borders are zoomed and cropped sections from the overview panels (cropped regions are indicated by the small dashed rectangle in the overview panels). The dashed scale bars in the cropped panels represent 20 µm. IHC was done on consecutive sections for the atg7−/− samples and each display two large, not recombined (i.e., Atg7wt/wt) regions of pancreatic acinar tissue that are marked as (*, **) and encircled in red. In addition two pancreatic islets (#, ##) are also marked and encircled which act as landmark structures to enable clear alignment of sequential sections. Antigen retrieval for LC3 was undertaken in Tris-EDTA, pH 9 for LC3 and in 10 mM Na Citrate buffer, pH 6 for ATG7, SQSTM1/p62 and ubiquitin.
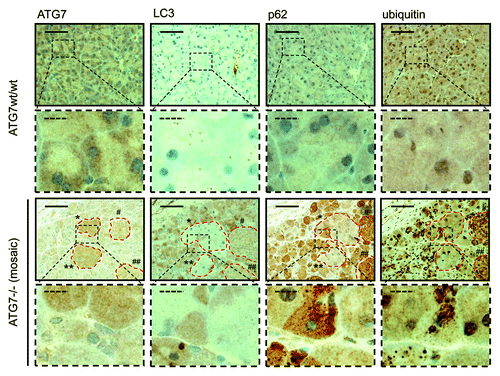
Figure 3. Detection of a punctate LC3-staining pattern in murine wild-type liver and uniform staining in atg7−/− tissue. Immunohistochemical staining for ATG7 and LC3 in murine wild-type liver (top two rows) and consecutive liver sections with mosaic deletion of ATG7 (atg7−/−, bottom two rows). Panels with straight borders are complete, i.e., not cropped and representative overview images taken at 40x magnification with a scale bar (straight) that represents 100 µm. Panels with dashed borders are cropped sections from the overview panels (cropped regions are indicated by the small dashed rectangle). The dashed scale bars in the zoomed and cropped panels represent 20 µm. IHC was done on consecutive sections for the atg7−/− samples and a landmark region with mostly recombined (i.e. ATG7-deficient) cells is encircled with red dashes. Antigen retrieval for LC3 was undertaken in Tris-EDTA, pH 9 and at 10 mM Na Citrate buffer, pH 6 for ATG7.
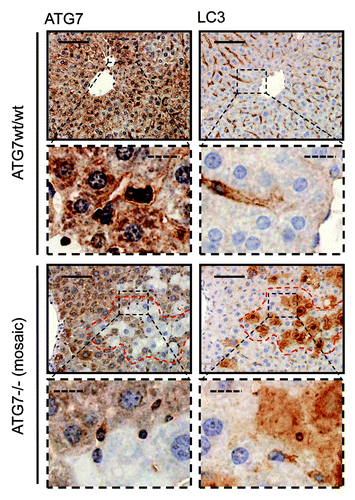
Figure 4. LC3 staining reveals quantitative differences in endogenous LC3 puncta formation. Representative immunohistochemical staining for LC3 in murine pancreas (A) and liver (B) from control mice (i.e., with access to food ad libitum) and mice starved for 14 h (but with free access to water). The panels are representative crops from images taken at 40x magnification and the dashed scale bars represent 20 µm. Box plots are used to display the quantification of LC3-puncta/cell for each condition. “*” indicates statistical significance (p < 0.05, Mann-Whitney). LC3-positive puncta of at least 150 cells (exocrine pancreas or hepatocytes respectively) from each of three mice in every group were counted. Antigen retrieval for LC3 was undertaken in Tris-EDTA, pH 9.
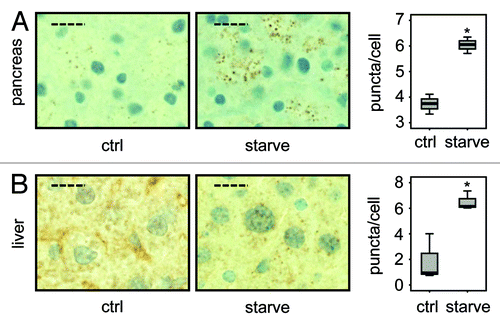
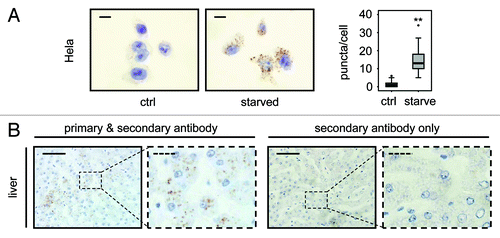
Figure 5. LC3 staining allows quantification of endogenous LC3 puncta in human cells and detects LC3 positive puncta in human tissue. (A) Representative immunohistochemical staining for LC3 in HeLa cells that were grown in replete medium or starved for 2 h in EBSS. Images were taken at 100x magnification and the scale bars represent 20 µm. Box plots are used to display the quantification of LC3-puncta/cell for each condition. “**” indicates statistical significance (p < 0.01, Mann-Whitney). LC3-positive puncta of 50 cells from each group were counted. (B) Human liver stained with LC3 antibody (left two panels) and without primary antibody as control (right two panels). Panels with straight borders are complete, i.e., not cropped and representative overview images taken at 40x magnification with a scale bar (straight) that represents 100 µm. Panels with dashed borders are zoomed and cropped sections from the overview panels (cropped regions are indicated by the small dashed rectangle in the overview panels). The dashed scale bars in the cropped panels represent 20 µm. Antigen retrieval for LC3 was undertaken in Tris-EDTA, pH 9.