Figures & data
Figure 1. Autophagy modulation rescues aged cells from SNCA-induced toxicity. Chronological life span of wild-type cells expressing SNCA WT, under the control of a Tet-On promoter, or harboring the vector control. SNCA WT expression was induced at (A) exponential, (B) diauxic or (C) stationary growth phases and cell viability was measured at 2–3 d intervals. SNCA WT levels in wild-type cells in which expression was induced at the different growth phases, (D) exponential, (E) diauxic or (F) stationary. (G) Chronological life span of stationary wild-type cells expressing SNCA WT in the presence or absence of chloroquine (CQ), an inhibitor of autophagy. The data represent mean ± SEM of three biological independent replicas. Significance of the chronological life span curves was determined by two-way ANOVA (***p < 0.001).
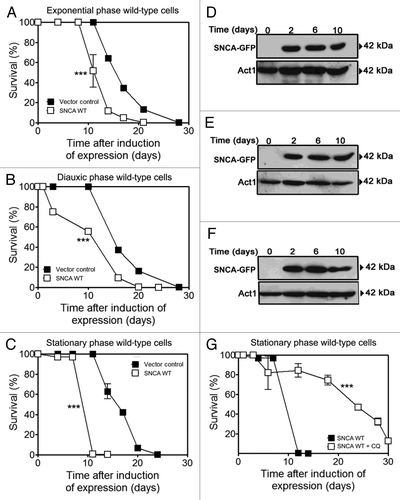
Figure 2. SNCA-induced toxicity in aged cells is associated with selective degradation of mitochondria. (A) Chronological life span and SNCA levels of wild-type cells expressing the vector control, the SNCA WT, the A53T or the A30P mutant under the control of the GAL1 promoter, a high toxicity model. (B and D) ATG8 and ATG32 mRNA relative expression levels at 0, 12 and 24 h after SNCA-induced expression. Three reference genes [ACT1 (actin), PDA1 (α subunit of pyruvate dehydrogenase) and TDH2 (isoform 2 of glyceraldehyde-3-phosphate dehydrogenase)] were used as internal standards and for the normalization of mRNA expression levels. (C) The splicing activation of the HAC1 mRNA at 0, 12 and 24 h after SNCA-induced expression. (E) Chronological life span and SNCA levels of wild-type cells expressing the vector control, the SNCA WT, the A53T or the A30P mutant under the control of the constitutive TPI1 promoter, a moderate toxicity model. The data represent mean ± SEM of six biological independent replicas. Cell viability was measured at 2–3 d intervals beginning at the day that cultures achieved stationary phase (day 0) and is expressed as % survival compared with survival at day 0 (100%). Autophagic and mitophagic activity were measured through the alkaline phosphatase assay that was performed in the wild-type cells expressing the vector control, the SNCA WT, the A53T or the A30P mutant and co-harboring a plasmid expressing the inactive Pho8 proenzyme targeted to (F) the cytosol or (G) to the mitochondrial matrix. The error bars represent the standard error of the mean (SEM). Significance of the data was determined by two-way ANOVA (*p < 0.05; **p < 0.01; ***p < 0.001).
![Figure 2. SNCA-induced toxicity in aged cells is associated with selective degradation of mitochondria. (A) Chronological life span and SNCA levels of wild-type cells expressing the vector control, the SNCA WT, the A53T or the A30P mutant under the control of the GAL1 promoter, a high toxicity model. (B and D) ATG8 and ATG32 mRNA relative expression levels at 0, 12 and 24 h after SNCA-induced expression. Three reference genes [ACT1 (actin), PDA1 (α subunit of pyruvate dehydrogenase) and TDH2 (isoform 2 of glyceraldehyde-3-phosphate dehydrogenase)] were used as internal standards and for the normalization of mRNA expression levels. (C) The splicing activation of the HAC1 mRNA at 0, 12 and 24 h after SNCA-induced expression. (E) Chronological life span and SNCA levels of wild-type cells expressing the vector control, the SNCA WT, the A53T or the A30P mutant under the control of the constitutive TPI1 promoter, a moderate toxicity model. The data represent mean ± SEM of six biological independent replicas. Cell viability was measured at 2–3 d intervals beginning at the day that cultures achieved stationary phase (day 0) and is expressed as % survival compared with survival at day 0 (100%). Autophagic and mitophagic activity were measured through the alkaline phosphatase assay that was performed in the wild-type cells expressing the vector control, the SNCA WT, the A53T or the A30P mutant and co-harboring a plasmid expressing the inactive Pho8 proenzyme targeted to (F) the cytosol or (G) to the mitochondrial matrix. The error bars represent the standard error of the mean (SEM). Significance of the data was determined by two-way ANOVA (*p < 0.05; **p < 0.01; ***p < 0.001).](/cms/asset/f6072eec-3535-45c8-83e9-fae538abadc9/kaup_a_10921275_f0002.gif)
Figure 3. SNCA-induced toxicity is mitophagy dependent. (A and B) Chronological life span and (C and D) SNCA levels of atg11Δ and atg32Δ cells, respectively, expressing the vector control, the SNCA WT, the A53T or the A30P mutant under the control of the constitutive TPI1 promoter. Cell viability was measured at 2–3 d intervals beginning at the day that cultures achieved stationary phase (day 0) and is expressed as % survival compared with survival at day 0 (100%). The data represents mean ± SEM of six biological independent replicas. (E and F) Mean (50% survival) and maximum (10% survival) chronological life spans were determined from curve fitting of the survival data (A and B) (from pair matched, pooled experiments) with the statistical software Prism (GraphPad Software). Significance was determined between wild-type cells and atg11Δ or atg32Δ cells expressing vector control or SNCA variants (*). The significance determined between cells expressing the vector control or SNCA variants within each strain (wild-type, atg11Δ or atg32Δ) was also determined (#). The alkaline phosphatase assay was performed to assess (G and H) autophagy or (I and J) mitophagy in atg11Δ or atg32Δ cells, respectively. The error bars represent the standard error of the mean (SEM). Significance of the data was determined by two-way ANOVA (#p < 0.05; *p < 0.05; **p < 0.01; ***p < 0.001).
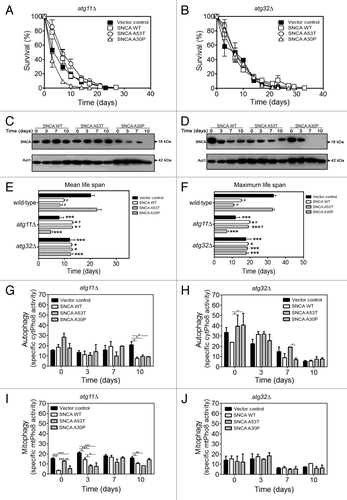
Figure 4. Mitophagy is involved in the SNCA-induced toxicity. Wild-type, atg11Δ and atg32Δ cells expressing, (A) mtDsRed and GFP-Atg8 and (B) SNCA WT were analyzed for mitophagy and autophagy by confocal fluorescence microscopy. SNCA WT was analyzed after incubation with antibody against SNCA as the primary and the Pacific Blue conjugated goat anti-rabbit secondary antibody. Single confocal planes are shown. Scale bars: 5 µm. (C) Foci number was quantified in wild-type, atg11Δ and atg32Δ cells expressing SNCA WT. The error bars represent the standard error of the mean (SEM).
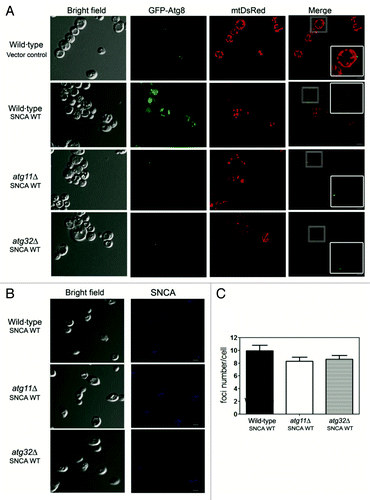
Figure 5. Superoxide anion accumulation under SNCA-induced toxicity is decreased in cells with impaired mitophagy. During chronological life span of (A) wild-type, (B) atg11Δ and (C) atg32Δ cells, expressing the vector control, the SNCA WT, the A53T or the A30P mutant forms under the control of the constitutive TPI1 promoter, the accumulation of superoxide anion was evaluated by flow cytometry, using the fluorescent probe dihydroethidium (DHE). The error bars represent the standard error of the mean (SEM). (D–G) Superoxide dismutase activity was analyzed in native gels. Bands corresponding to Sod1 activity of cells expressing vector control (D) or SNCA A53T mutant (F) and Sod2 activity of cells expressing vector control (E) or SNCA A53T mutant (G) were measured with a densitometer and the relative intensities calculated (Quantity One software, BioRad). Values indicate mean ± SEM from three independent experiments. Significance of the values between yeast strains was determined by two-way ANOVA (*p < 0.05; **p < 0.01; ***p < 0.001).
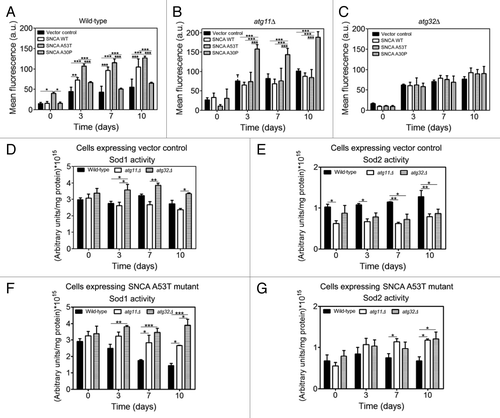
Figure 6. Sir2 mediates autophagy/mitophagy induction under SNCA-induced toxicity. (A) Chronological life span and (B) SNCA levels of sir2Δ cells expressing the vector control, the SNCA WT, the A53T or the A30P mutant under the control of the constitutive TPI1 promoter. Cell viability was measured at 2–3 d intervals beginning at the day that cultures achieved stationary phase (day 0) and is expressed as % survival compared with survival at day 0 (100%). The alkaline phosphatase assay was performed to assess (C) autophagy or (D) mitophagy, respectively. (E) sir2Δ cells expressing mtDsRed and GFP-Atg8 were analyzed for mitophagy and autophagy by confocal fluorescence microscopy. Single confocal planes are shown. Scale bars: 5 µm. The error bars represent the standard error of the mean (SEM). (F–I) Relative ATG8 and ATG32 mRNA levels in stationary phase wild-type and sir2Δ cells expressing SNCA at day 0, 3, 7 and 10. Three reference genes [ACT1 (actin), PDA1 (α subunit of pyruvate dehydrogenase) and TDH2 (isoform 2 of glyceraldehyde-3-phosphate dehydrogenase)] were used as internal standards and for the normalization of mRNA expression levels. Significance of the data was determined by two-way ANOVA (*p < 0.05; **p < 0.01; ***p < 0.001).
![Figure 6. Sir2 mediates autophagy/mitophagy induction under SNCA-induced toxicity. (A) Chronological life span and (B) SNCA levels of sir2Δ cells expressing the vector control, the SNCA WT, the A53T or the A30P mutant under the control of the constitutive TPI1 promoter. Cell viability was measured at 2–3 d intervals beginning at the day that cultures achieved stationary phase (day 0) and is expressed as % survival compared with survival at day 0 (100%). The alkaline phosphatase assay was performed to assess (C) autophagy or (D) mitophagy, respectively. (E) sir2Δ cells expressing mtDsRed and GFP-Atg8 were analyzed for mitophagy and autophagy by confocal fluorescence microscopy. Single confocal planes are shown. Scale bars: 5 µm. The error bars represent the standard error of the mean (SEM). (F–I) Relative ATG8 and ATG32 mRNA levels in stationary phase wild-type and sir2Δ cells expressing SNCA at day 0, 3, 7 and 10. Three reference genes [ACT1 (actin), PDA1 (α subunit of pyruvate dehydrogenase) and TDH2 (isoform 2 of glyceraldehyde-3-phosphate dehydrogenase)] were used as internal standards and for the normalization of mRNA expression levels. Significance of the data was determined by two-way ANOVA (*p < 0.05; **p < 0.01; ***p < 0.001).](/cms/asset/dea99e17-be2c-4fc5-9455-264a77a97b70/kaup_a_10921275_f0006.gif)