Figures & data
Figure 1. Ethanol exposure increased LC3 content, decreased proteasome activity and enhanced AV content in VL-17A cells. (A) Mean densitometric ratios of LC3-I and LC3-II, each to β-actin (ACTB) in cells treated 24 h with the indicated doses of ethanol. (B) Proteasome activity in cells treated 24 h with the indicated doses of ethanol. Data are mean values of the LC3 protein bands from western blots (A) proteasome chymotrypsin-like activity (B) from quadruplicate culture flasks of each treatment group. (C) AV content in VL-17A cells: Cells were exposed to the conditions indicated in the figure for 24 h. After treatment, the cells were fixed in 4% paraformaldehyde and immuno-stained for AVs using anti LC3B. Images were captured by confocal microscopy. Green puncta are AVs. Nuclei stained with DAPI are shown in blue. Microscopy data (C) indicated are mean values from 400 to 1100 cell images. (D) AV numbers, size and its proportion to total cell area (fractional volume) in cells from (C). Letter superscripts or letters in the figures that are different from each other indicate that the data are significantly different from each other. Data with the same letter are not significantly different.
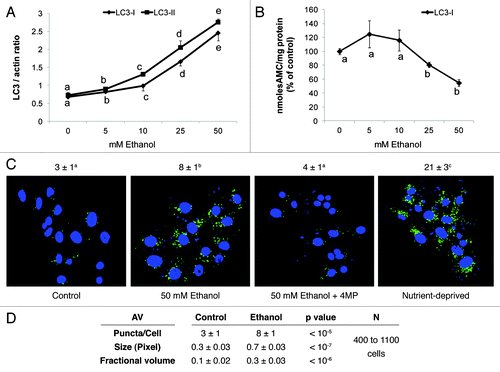
Figure 2. Ethanol exposure enhanced LC3B mRNA and inhibited MTORC1. (A) LC3B mRNA levels in VL-17A cells after 12 and 24 h exposure to ethanol or nutrient deprivation. (B) Ethanol effect on RPS6K in VL-17A cells. Mean densitometric ratios of pRPS6K/RPS6K in cells treated 24 h with zero or 50 mM ethanol. Data are mean values (± SEM) from quadruplicate samples. Letters that are different from each other indicate that the data are significantly different from each other. Data with the same letter are not significantly different.
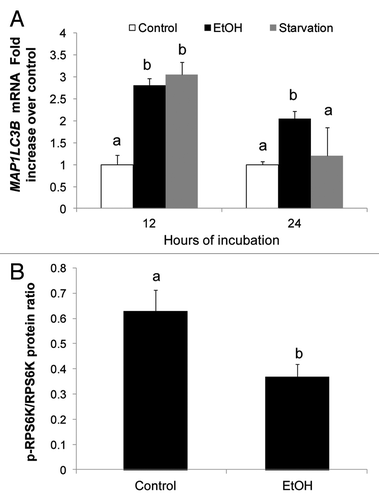
Figure 3. Ethanol exposure influenced LC3-II and SQSTM1 flux, mimicked the effects of nocodazole, and decreased AV-lysosome colocalization. (A) Flux measurements of LC3-II in VL-17A cells treated with or without 50 mM ethanol in the presence or absence of bafilomycin A1 (added during the last 4 h) and quantified as described in Methods. (B) Quantification of SQSTM1 under the same conditions as described in (A). (C) LC3-II levels in VL-17A cells after 24 h exposure to 50 mM ethanol, 10 µM nocodazole or 100 nM rapamycin. (D) SQSTM1 levels in VL-17A cells after 24 h exposure to 50 mM ethanol, 10 µM nocodazole or 100 nM rapamycin. Data are mean values (± SEM) from quadruplicate flasks per treatment. (E) AVs, lysosomes and AV-lysosome colocalization in VL-17A cells. Cells were transduced with adenovirus encoding GFP-LC3 and exposed to zero or 50 mM ethanol for 24 h. Images were captured by confocal microscopy. Green puncta are AVs and red dots are lysosomes. Nuclei stained with Hoechst are shown in blue. (F) Cells exposed to zero or 50 mM ethanol for 24 h and immunostained for AVs and LAMP1. Green puncta are AVs and red dots are LAMP1. Nuclei stained with DAPI are shown in blue. (G) Lysosome (LAMP1) number, size and its proportion to total cell area (Fractional volume) in cells from (F). Numerical data given above each image are mean AV, or lysosome, (represented as LysoTracker Red or LAMP1) puncta numbers or co-localized AVs with lysosomes (merged images) per nucleus obtained from 600 to 800 cell images per treatment group. Letter superscripts that are different from each other indicate that the data are significantly different from each other between the treatment groups. Data with the same letter superscript are not significantly different.
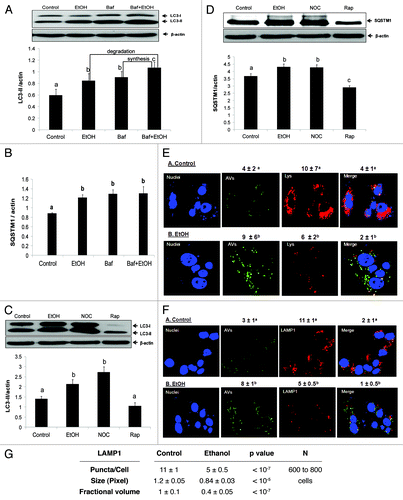
Figure 4. Ethanol dose-dependently affected cathepsin activity. (A) CTSB-specific activity, and (B) CTSL-specific activity in VL-17A cells after 24 h treatment with the indicated ethanol concentrations. Data are mean values (± SEM) from quadruplicate flasks per treatment. Letters that are different from each other indicate that the data are significantly different from each other. Data with the same letter are not significantly different.
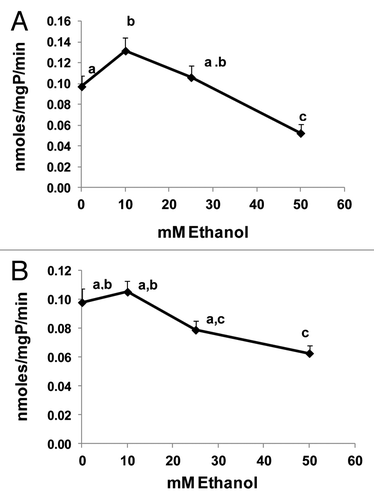
Figure 5. Ethanol exposure differentially affected LC3-II levels, MDA and acetaldehyde generation in four Hep G2 cell lines that differentially express ethanol metabolizing enzymes. (A) Representative western blot showing ADH and CYP2E1 protein phenotypes in the four cell lines indicated. (B) Representative western blot and mean densitometric ratios of LC3-II to ACTB in the four Hep G2 cell lines after 24 h exposure to zero or 50 mM ethanol. (C) Malondialdehyde levels in the four cell lines after exposure to zero or 50 mM ethanol. (D) Mean acetaldehyde levels in media from the Hep G2 cells lines after 24 h exposure to 50 mM ethanol. Data are mean values (± SEM) from 4 to16 culture flasks of each treatment group. Letters that are different from each other indicate that the data are significantly different from each other. Data with the same letter are not significantly different.
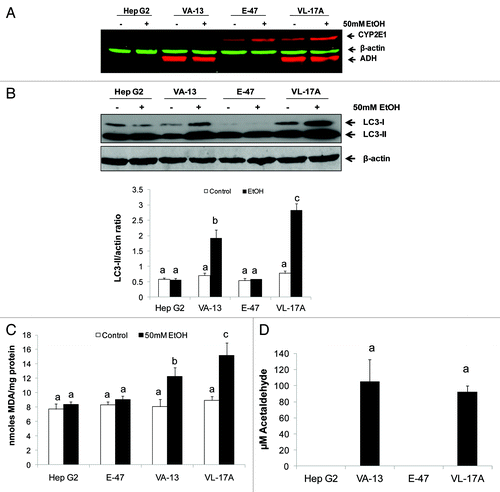
Figure 6. Ethanol and acetaldehyde exposures enhanced LC3-II levels in ethanol-metabolizing and nonmetabolizing Hep G2 cells. (A) LC3-II content after 24 h treatment with 50 mM ethanol with or without 5 mM 4-methylpyrazole (4MP), 5mM glutathione ethyl ester (GSH-EE), 100 µM chlormethiazole (CMZ) or 20 µM trolox. (B) LC3II content after 24 h exposure of VL-17A cells or VA-13 cells to 50 mM ethanol or to 100 µM acetaldehyde. (C) LC3-II levels in Hep G2 cells after treatment for 1 h with 300 µM acetaldehyde. Data are mean densitometric ratios of LC3-II to ACTB from quadruplicate culture flasks. Letters that are different from each other indicate that the data are significantly different from each other. Data with the same letter are not significantly different.
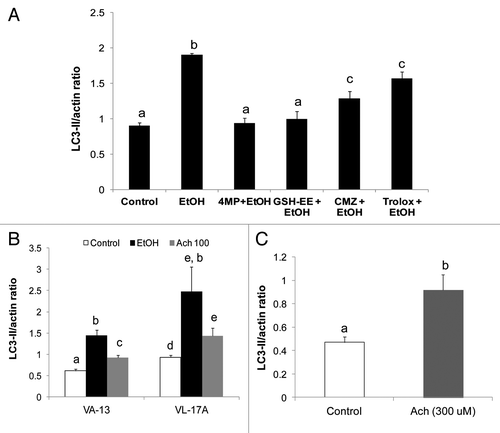
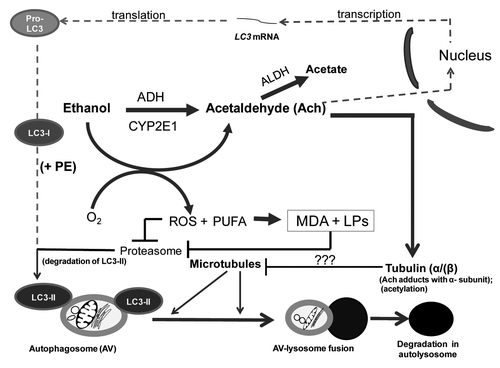
Figure 7. Multilevel regulation of autophagosome content by ethanol oxidation liver cells. During acute (or early) ethanol administration, metabolically-derived acetaldehyde (Ach) enhances AV formation in liver cells by increasing the level of LC3B mRNA (presumably by enhanced transcription). LC3B mRNA is then translated into pro-LC3, which matures to LC3-I. The latter is lipidated with phosphatidylethanolamine (PE) to form LC3-II, which attaches to the AV membrane. The AV is trafficked by microtubules to the lysosome for degradation. During habitual (chronic) ethanol exposure, CYP2E1 is induced, generating MDA and other lipid peroxides (LPs), which inhibit proteasome activity and stabilize LC3-II from degradation. Acetaldehyde (Ach) generated by ethanol oxidation forms adducts with proteins, including the α tubulin subunit.Citation19 We hypothesize that formation of Ach-α-tubulin adducts or tubulin acetylation block or stabilize the polymerization of microtubules (see “???” in lower half of figure). Either change will prevent the fusion of AVs with lysosomes, thereby causing ethanol-induced proteopathy and steatosis in liver cells.