Figures & data
Figure 1. The cytosolic sequence of LAMP2C interacts with RNA-binding proteins. (A) Identity levels of the cytosolic sequences of fly and nematode LAMPs to that of human LAMP2s. (B) A schematic representation of biotin-conjugated peptides. (C) Proteins that interact with the cytosolic sequences of LAMP2s were analyzed by pull-down assay followed by LC-MS/MS analysis of LAMP2C peptide-interacting proteins. (D) Expression levels of Lamp2c mRNA in mouse tissues. Values are the means ± SE (n = 3 to 4). (E) Pull-down assay and LC-MS/MS analysis of LAMP2C peptide-interacting proteins from mouse brain. The distribution of the identified proteins is shown.
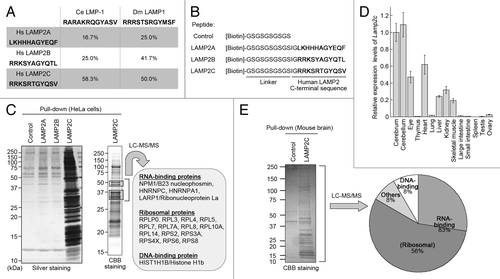
Figure 2. The cytosolic sequence of LAMP2C directly interacts with RNA. (A) Protein interactions of LAMP2C peptide were analyzed by pull-down assay using brain lysates preincubated with or without RNase A. (B) Pull-down assay using lysates of HeLa cells transfected with the indicated constructs. (C) A pull-down assay was performed using brain lysate, and RNA was detected with EtBr. The intense signal in the input lane (*) is presumably degraded RNA in the brain lysate. (D) Interactions of purified total RNA with cytosolic sequence of LAMP2C. Amounts of RNA remaining in the flow-through fraction were quantified by measuring OD260 (n = 4). (E) Interactions of purified total RNA with cytosolic sequences of nematode and fly LAMPs.
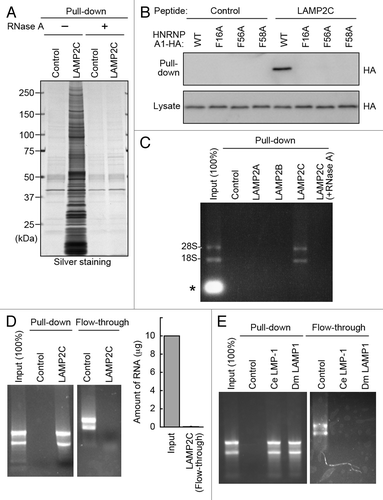
Figure 3. Uptake and degradation of RNA by isolated lysosomes. (A and B) Uptake of RNA by isolated lysosomes. Isolated lysosomes were incubated with purified total RNA (5 μg) in the presence or absence of ATP (energy regeneration system) and/or HSPA8. Levels of RNA remaining in solution outside of lysosomes (A) and levels of RNA resistant to exogenous RNase A (B) were analyzed (n = 3). (C) Lysosomes were incubated with 10 μg of RNA in the presence or absence of ATP. At the indicated times, the levels of RNA remaining outside of lysosomes were analyzed (n = 3). (D) Degradation of RNA by isolated lysosomes. Lysosomes and RNA were incubated with or without ATP. Total levels of RNA in the incubated samples were analyzed (n = 3). (E) Immunogold labeling of RNA in isolated lysosomes incubated with ATP and RNA. Immunogold labeling was performed using an anti-rRNA antibody followed by anti-mouse IgG coupled with 10 nm of gold particles. Gold particles were observed in the lysosomes.
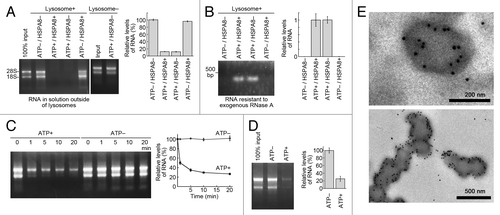
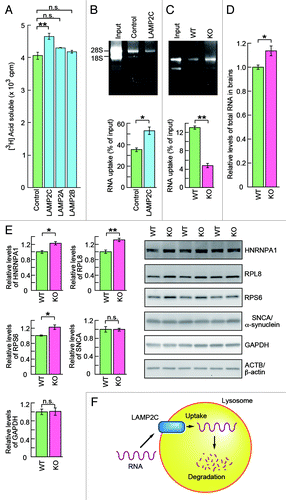
Figure 4. LAMP2C mediates lysosomal degradation of RNA. (A) RNA turnover in HeLa cells transfected with each LAMP2 isoform or empty vector (n = 3). (B and C) Uptake of RNA into lysosomes isolated from HeLa cells transfected with LAMP2C or empty vector (n = 3) (B). Uptake of RNA into lysosomes isolated from the brains of wild-type (WT) and LAMP2 knockout (KO) mice (n = 3) (C). Levels of RNA uptake were measured by subtracting the levels of RNA remaining in solution outside of lysosomes from the levels of input RNA. (D) Relative levels of total RNA in the brains of WT and LAMP2 KO mice (KO: n = 3, WT: n = 4). (E) Levels of RBPs and CMA substrate proteins in the brains of WT and LAMP2 KO mice were analyzed by immunoblotting (n = 3/group). (F) Model for the possible mechanism of RNautophagy. LAMP2C on the lysosomal membrane recognizes RNA, and the RNA is then imported into the lysosomal lumen in an ATP-dependent manner, followed by its degradation. The requirement for ATP in RNautophagy suggests an involvement of ATPases such as RNA helicases in the process on the lysosomal membrane.