Figures & data
Figure 1. Loss of ULK1 and ULK2 inhibits autophagy following amino acid starvation. (A) Wild-type (WT), Ulk1 knockout (ULK1KO), Ulk2 knockout (ULK2KO) and two independently derived Ulk1/Ulk2 double knockout (DKO) MEF lines were treated with full-nutrient medium (F), EBSS (-AA) or EBSS containing 50 nM bafilomycin A1 (Baf) for 2 h. Resolved cell lysates were immunoblotted with antibodies against ULK1, actin and LC3 (Nanotools 5F10 monoclonal). “*” indicates nonspecific band on ULK1 immunoblot. MEF lines were immortalized with SV40 T antigen. Below: LC3-II/LC3-I signals were plotted as average ± SEM, (n = 4) using 2 different y-axis scales. Two-tailed paired t-tests were performed relative to values of WT MEFs under the same condition: *p < 0.04; **p < 0.005; ap = 0.14; bp = 0.056; cp = 0.12. (B) Total RNA was extracted from MEF lines described in (A) and analyzed for Ulk1 and Ulk2 transcript levels using quantitative RT-PCR. Plots show Ulk1 or Ulk2 levels normalized to Actin transcript levels (average ± SEM, (n = 3), values relative to WT cells). (C) WT and DKO MEF lines were treated as in (A) to full-nutrient medium or EBSS for 2 h. Resolved cell lysates were immunoblotted with antibodies against phospho-(Ser79) acetyl CoA carboxylase (P-ACC), phospho-(Ser240/244) ribosomal protein S6 (P-RPS6); actin and LC3 (5F10 monoclonal antibody). (D) MEF lines were treated to full-nutrient medium (F) or glucose starvation (−Glc) for 16 h before lysis and immunoblot analysis as in (C). (E) MEF lines were steady-state labeled overnight with C14-valine, chased for 24 h, and analyzed for protein degradation over 2 h of amino acid starvation. Plot shows average ± SEM (n = 3). Two-tailed paired t-tests were performed relative to WT MEFs: *p < 0.02; **p < 0.01. (F) WT and Ulk DKO#1 MEFs were treated as in (A) for 2 h and fixed for LC3 immunostaining. Shown are representative confocal images of −AA and −AA+Baf conditions. Scale bar: 10 μm. Plots show average puncta/cell ± SEM (each column: n = 22 to 39 cells per condition from two experiments). Two-tailed paired t-tests when compared with WT MEFs under the same condition: ***p < 0.0003; *p < 0.02. (G) Primary WT and DKO MEFs were subjected to starvation conditions as in (A) and lysed for immunoblot analysis of SQSTM1 and actin. Right: Plotted densitometry [SQSTM1 levels normalized to actin, shown as average ± SEM (n = 4)]. Two-tailed paired t-tests when compared with WT MEFs under same conditions: *p < 0.04, **p < 0.01.
![Figure 1. Loss of ULK1 and ULK2 inhibits autophagy following amino acid starvation. (A) Wild-type (WT), Ulk1 knockout (ULK1KO), Ulk2 knockout (ULK2KO) and two independently derived Ulk1/Ulk2 double knockout (DKO) MEF lines were treated with full-nutrient medium (F), EBSS (-AA) or EBSS containing 50 nM bafilomycin A1 (Baf) for 2 h. Resolved cell lysates were immunoblotted with antibodies against ULK1, actin and LC3 (Nanotools 5F10 monoclonal). “*” indicates nonspecific band on ULK1 immunoblot. MEF lines were immortalized with SV40 T antigen. Below: LC3-II/LC3-I signals were plotted as average ± SEM, (n = 4) using 2 different y-axis scales. Two-tailed paired t-tests were performed relative to values of WT MEFs under the same condition: *p < 0.04; **p < 0.005; ap = 0.14; bp = 0.056; cp = 0.12. (B) Total RNA was extracted from MEF lines described in (A) and analyzed for Ulk1 and Ulk2 transcript levels using quantitative RT-PCR. Plots show Ulk1 or Ulk2 levels normalized to Actin transcript levels (average ± SEM, (n = 3), values relative to WT cells). (C) WT and DKO MEF lines were treated as in (A) to full-nutrient medium or EBSS for 2 h. Resolved cell lysates were immunoblotted with antibodies against phospho-(Ser79) acetyl CoA carboxylase (P-ACC), phospho-(Ser240/244) ribosomal protein S6 (P-RPS6); actin and LC3 (5F10 monoclonal antibody). (D) MEF lines were treated to full-nutrient medium (F) or glucose starvation (−Glc) for 16 h before lysis and immunoblot analysis as in (C). (E) MEF lines were steady-state labeled overnight with C14-valine, chased for 24 h, and analyzed for protein degradation over 2 h of amino acid starvation. Plot shows average ± SEM (n = 3). Two-tailed paired t-tests were performed relative to WT MEFs: *p < 0.02; **p < 0.01. (F) WT and Ulk DKO#1 MEFs were treated as in (A) for 2 h and fixed for LC3 immunostaining. Shown are representative confocal images of −AA and −AA+Baf conditions. Scale bar: 10 μm. Plots show average puncta/cell ± SEM (each column: n = 22 to 39 cells per condition from two experiments). Two-tailed paired t-tests when compared with WT MEFs under the same condition: ***p < 0.0003; *p < 0.02. (G) Primary WT and DKO MEFs were subjected to starvation conditions as in (A) and lysed for immunoblot analysis of SQSTM1 and actin. Right: Plotted densitometry [SQSTM1 levels normalized to actin, shown as average ± SEM (n = 4)]. Two-tailed paired t-tests when compared with WT MEFs under same conditions: *p < 0.04, **p < 0.01.](/cms/asset/ca9901af-a0b6-4fab-969b-9d1b8a5eb055/kaup_a_10923066_f0001.gif)
Figure 2. ULK1 and ULK2 are both required for WIPI2 spot formation following amino acid starvation. (A) WT, ULK1KO, ULK2KO and DKO primary MEFs were treated with either full nutrient-medium (F) or EBSS (−AA) for 2 h before fixation. Cells were then stained with an antibody against WIPI2. (B) In 10 fields per condition (two independent experiments) Hoechst-stained nuclei (cell number) and WIPI2 puncta were quantified (WIPI2 puncta/cell ± SEM) (n = 20 fields). Differences between F and −AA are indicated by ****p < 0.0001 by one-way ANOVA with Bonferroni post hoc test, n.s. non-significant. (C) Primary MEF lines were treated as in (A). Cell lysates were immunoblotted for ULK1, WIPI1, WIPI2 and actin. (D) WT and DKO#1 immortalized cells stably expressing GFP-ZFYVE1 were treated to full-nutrient medium (F) or EBSS (−AA) for 2 h before fixation. (A and D) Scale bars: 10 μm. Plot shows puncta/cell (average ± SEM for n = 29 to 37 cells per condition from two experiments). Two-tailed paired t-test comparing to WT MEFs under same conditions: ***p < 0.0001.
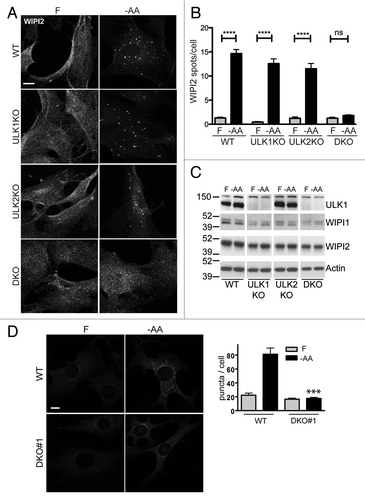
Figure 3. ATG13 knockdown phenocopies Ulk1/Ulk2 double knockout for the LC3 lipidation pathway following amino acid starvation. (A) WT and DKO primary MEFs were transfected with either RISC-Free control or mouse Atg13-targeting siRNA. Following knockdown, cells were treated with either full-nutrient medium (F), EBSS (−AA) or EBSS containing bafilomycin A1 (−AA+Baf) (100 nM) for 2 h. Resolved cell lysates were immunoblotted with antibodies against ULK1, ATG13, actin and LC3 (Abcam polyclonal). (B) Average LC3-II/actin densitometry ± SEM (n = 4). Two-tailed paired t-tests were performed relative to WT-RISC-Free siRNA MEFs under the same conditions: ap = 0.067; bp = 0.068; cp = 0.089.
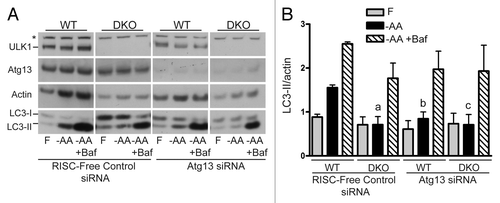
Figure 4. ATG13 knockdown phenocopies Ulk1/Ulk2 double knockout on the formation of WIPI2-positive autophagosomal membranes following amino acid starvation. WT, ULK1KO, ULK2KO and DKO primary MEFs were transfected with either RISC-Free control or mouse Atg13-targeting siRNA. Following knockdown, cells were treated with either full-nutrient medium (F) or EBSS (−AA) for 2 h before fixation. Cells were stained with an antibody against WIPI2. Scale bar:10 μm.
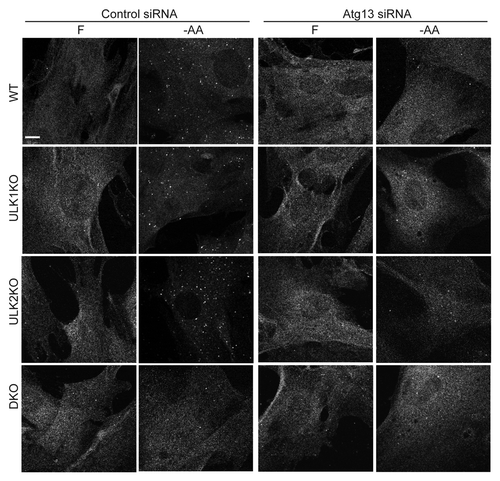
Figure 5. Combined Ulk1 and Ulk2 knockout reduces the LC3 lipidation response to glucose starvation. (A) WT or DKO primary MEFs were treated to either full-nutrient medium (F), glucose-free conditions (−Glc), or glucose-free medium + bafilomycin A1 (−Glc+Baf) (100 nM) for 24 h. Resolved cell lysates were immunoblotted with antibodies against ULK1, actin and LC3 (Abcam polyclonal). “*” indicates nonspecific band. (B) LC3-II and actin signals were quantified by densitometry. Results are shown as average ± SEM (n = 4). Two-tailed paired t-tests were performed relative to WT MEFs under the same treatment: *p < 0.02.
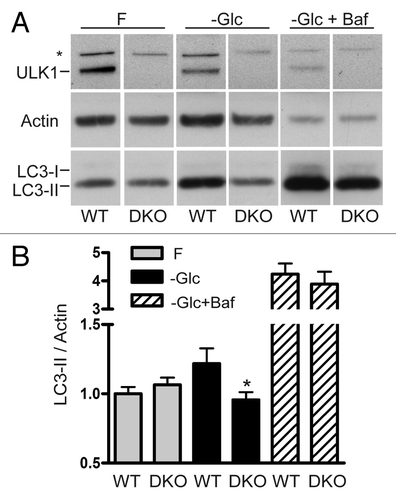
Figure 6. Glucose starvation does not stimulate production of WIPI2-containing membranes. WT, ULK1KO, ULK2KO and DKO primary MEFs were treated with either full-nutrient medium (Fed) or glucose-free medium (glucose starved) for 24 h then fixed. Cells were stained with an antibody against WIPI2. Scale bar:10 μm.
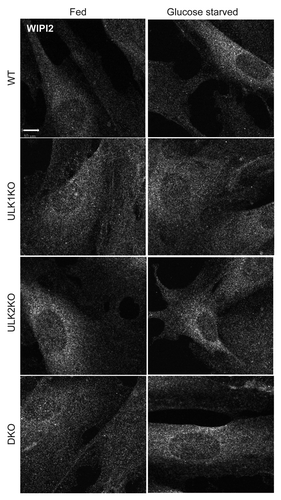
Figure 7. LC3 lipidation and autophagosome formation following glucose starvation are PtdIns3P-independent. (A) WT primary MEFs were treated with full-nutrient medium (F), EBSS (−AA), or EBSS containing 100 nM wortmannin (−AA+Wm) for 2 h. Alternatively, cells were incubated in glucose-free medium (−Glc) or glucose-free medium containing 100 nM wortmannin (−Glc+Wm) for 24 h. Resolved cells lysates were immunoblotted with antibodies for LC3 (Abcam polyclonal) and actin. Results are shown as average LC3-II/actin ± SEM (n = 4). Two-tailed paired t-tests were performed relative to equivalent treatment condition without wortmannin: *p < 0.04. (B) WT (immortalized) MEFs were treated with full-nutrient medium (F) or EBSS (−AA) for 2 h. Alternatively, cells were incubated in glucose-free medium (−Glc) for 16 h. Where indicated, incubations included 100 nM wortmannin (+Wm). Cells were fixed after treatments for LC3 immunostaining. Shown are representative confocal images of −AA and −Glc conditions ± wortmannin. Scale bar: 10 μm. Plot shows puncta/cell distributions for each condition. Each column represents quantification of one cell (40–60 cells/condition taken from two experiments). Shown are percentages of cells within each group containing 15 (or more) puncta/cell.
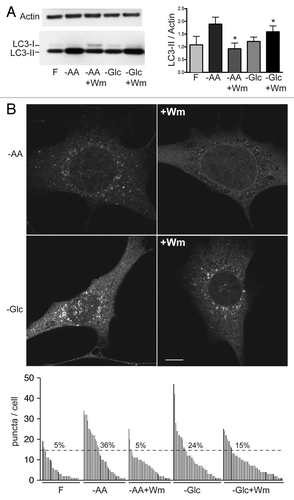
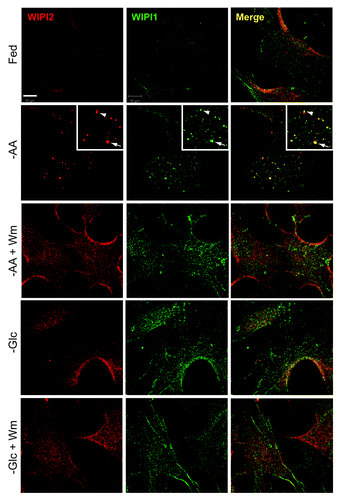
Figure 8. Absence of WIPI1- and WIPI2-positive autophagosomal membranes following glucose starvation. WT primary MEFs were incubated in full-nutrient medium (Fed), EBSS (−AA), or EBSS with 100 nM wortmannin (−AA+Wm) for 2 h. Alternatively, cells were glucose starved without or with 100 nM wortmannin (−Glc, −Glc+Wm) for 24 h before fixation. Cells were stained with antibodies against WIPI2 (red) and WIPI1 (green). Scale bar: 10 μm. Zoomed inset shows WIPI2 and WIPI1 colocalization following amino acid starvation (arrow: strong colocalization; arrowhead: partial).