Figures & data
Figure 1. Evaluation of the experimental error of the time-course measurement of ATG protein accumulation into punctate structures. MEFs stably coexpressing CFP–ATG5 and Venus–ATG5 (A), CFP-ATG16L1 and Venus–ATG5 (B), CFP–ATG5 and Venus–ZFYVE1 (C), or CFP–ZFYVE1 and Venus–ATG5 (D) were cultured in starvation medium. Time-lapse imaging was performed at 1 frame per 10 s, and selected images (0.5 min interval) are shown. The time course of the fluorescence intensity of the punctate signals shown in the images (left) was plotted in the graphs (right; percent maximum intensity). Dashed-line boxes indicate the video frames at the maximum (peak) intensities. Scale bar: 5 μm.
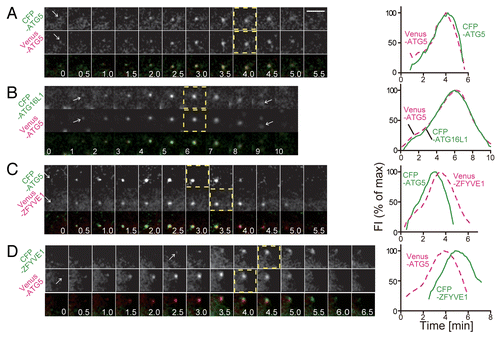
Figure 2. Time-course analysis of accumulation and disappearance of ATG proteins. MEFs stably coexpressing CFP–ATG5 and Venus–ULK1 (A), CFP–ATG5 and Venus–ATG14 (B), CFP–WIPI1 and Venus–ATG5 (C), CFP–WIPI1 and Venus–ZFYVE1 (D), CFP–LC3 and Venus–DCFP1 (E), or CFP–SQSTM1 and Venus–LC3 (F) were cultured in starvation medium. Time-lapse imaging was performed and analyzed as in . Selected images (1.0 min interval) are shown. Dashed-line boxes indicate the video frames at the maximum (peak) intensities. Scale bar: 5 μm.
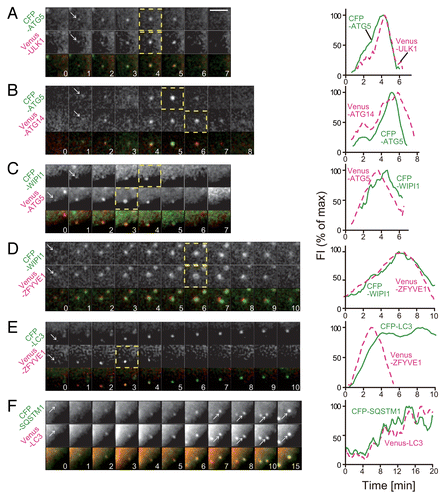
Figure 3. Lag of the peak time of accumulation between ATG proteins. (A) Average fluorescence intensity profiles of all data sets are plotted. The number of samples is indicated in each panel. (B) Histograms show the distribution for the time lag of the maximum peaks of fluorescence intensity profiles between the indicated pairs of CFP- and Venus-fused ATG proteins in (A). Black horizontal, and blue and red vertical bars indicate the standard deviation (sd), mean, and median, respectively. P values were obtained using the unpaired Student t-test between indicated pairs of proteins.
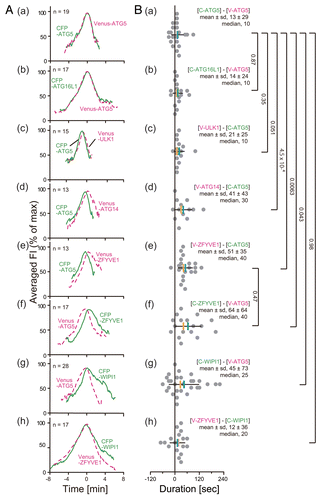
Figure 4. PtdIns3K is important for stable accumulation of ULK1 and ATG5. MEFs stably coexpressing CFP–ATG5 and Venus-ULK1 were cultured in starvation medium for 30 min (upper panels) and then 0.2 μM wortmannin (WM) was added (lower panels). The ULK1+ ATG5+ punctate structures were tracked for 5 min after the addition of wortmannin by time-lapse microscopy and were classified into the four groups: ULK1− ATG5− (arrows), ULK1+ ATG5− (arrowheads), ULK1− ATG5+ and ULK1+ ATG5+. Relative population of each group was shown in the table (right). Scale bar: 10 μm.
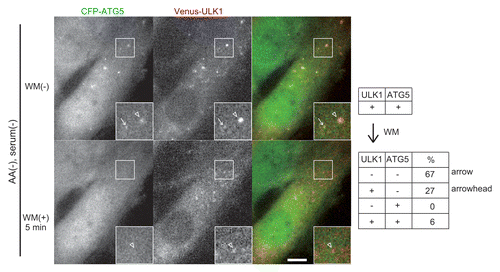
Figure 5. ATG9 vesicles are transiently recruited to the autophagosome formation site. (A) Structured illumination microscopy (SIM) imaging of MEFs stably expressing GFP–ATG9A cultured in starvation medium. Scale bar: 10 μm. (B) Time-lapse images of GFP–ULK1 and mRFP–ATG9A in starved MEFs were simultaneously produced at a rate of 160 ms/frame by conventional fluorescence microscopy using a fluorescence-split system. Selected images (1.6 s interval) are shown. Arrowheads indicate mRFP–ATG9A structures that are associated with a GFP–ULK1 punctate structure. Scale bar: 2 μm.
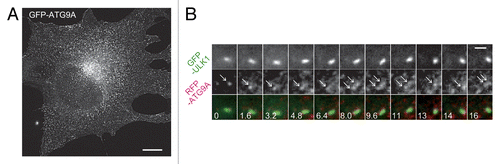
Figure 6. ULK1 is recruited to pre-existing VMP1-positive punctate structures. (A) SIM imaging of MEFs stably expressing VMP1–GFP cultured in normal medium. Scale bar: 10 μm. (B) Time-lapse imaging of VMP1–CFP and Venus–ULK1 in starved MEFs was performed at 1 frame per 10 s, and selected images (0.5 min interval) are shown. Arrows indicate VMP1–CFP and Venus–ULK1 puncta, which colocalize with each other for certain times. Scale bar: 5 μm.
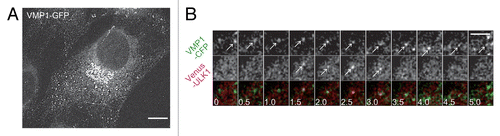
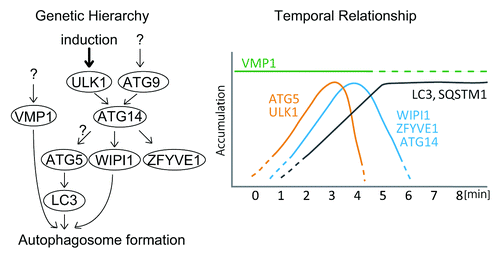
Figure 7. Schematic models of the genetic hierarchy and temporal relationship among mammalian ATG proteins. Genetic hierarchy of mammalian ATG proteins based on previous reportsCitation6-Citation8 (left) and their temporal relationship revealed by the current study (right) are shown.