Figures & data
Figure 1. Generation of inducible cell lines for deletion mutants of mouse ARF. (A) Schematic of the coding region of Arf. Exon 1β contains the binding sites for MDM2 and nucleophosmin/B23 (NPM). Exon 2, shared in an alternative reading frame with the sequence for the p16INK4a protein, is the site for the majority of tumor-derived mutations in CDKN2A. Internal initiation of translation at methionine M45 generates a short form of ARF, denoted smARF. The nucleolar localization sequence is denoted. (B) Western blot analysis of murine ARF deletion mutants in stably transfected clones of U2OS-ARF cell lines untreated or treated with 0.1 µg/mL doxycycline (doxy) for 24 h. Actin (ACTB) is included as a loading control. Bands for ARF and smARF are denoted; note that a small amount of smARF is generated in the wt ARF-induced cells (lane 2). (C) Immunofluorescence analysis of U2OS-ARF cells treated with 0.1 µg/mL doxycycline (+doxy) for 48 h, immunostained with antisera to murine ARF (green) and β-tubulin (TUBB, red), and stained with DAPI (blue).
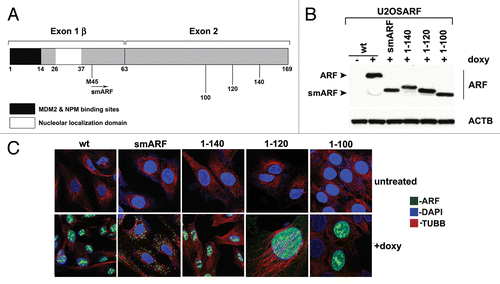
Figure 2. The mitochondrial localization sequence of murine p19ARF maps to amino acids 45 to 100; murine smARF shows enhanced localization to mitochondria. (A) Western blot analysis of whole cell lysate (wcl) vs. lysate from purified mitochondria (mito) isolated from U2OS-ARF cells following treatment with doxycycline for 24 h (doxy, 0.1 μg/ml). Lysates were probed for the mitochondrial protein HSPA9 (GRP75) and the nuclear/cytosolic protein PCNA as an assessment of purity. The bottom lanes depict ARF protein level in these fractions. The data depicted are representative of three independent experiments, for multiple clones for each mutant. (B) Immuno-electron microscopy using ARF antisera followed by protein G-Gold in U2OS-ARF cells following 24 h treatment with doxycycline. The arrowheads depict gold particles colocalizing with mitochondria. Scale bar: 500 nm. (C) Graphic analysis of the percent of mitochondria immunostained with ARF antisera in (B). Mitochondria with four or greater gold particles were considered positive, and at least 200 mitochondria were counted for each sample.
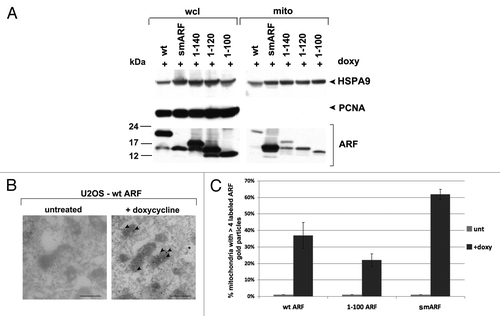
Figure 3. The murine ARF autophagy domain maps to amino acids 100 to 120. (A) Western blot analysis of U2OS-ARF cells containing the deletion mutants indicated, untreated or treated with doxycycline (doxy, 0.1 μg/ml) for the indicated time points (hours). Degradation of SQSTM1/p62 (SQSTM1) and accumulation of LC3-II (bottom LC3 band) are indicators of autophagy induction. Actin (ACTB) is included as a loading control. The data depicted are representative of three independent experiments, for multiple clones of each mutant. (B) Western analysis of the level of SQSTM1 and LC3-I and -II following cessation of autophagic flux with 10 mM NH4Cl. Note that whereas wt ARF induces increased LC3-II, above that which accumulates following NH4Cl (compare lanes 4 and 3), there is no increase in LC3-II in smARF-induced cells (compare lanes 8 and 7). (C) Electron microscopy of U2OS-ARF cells containing wild-type ARF (wt) or the deletion mutants indicated, untreated (unt) or treated with doxycycline (0.1 µg/mL) for 48 h. The arrowheads point to autophagic vesicles that appear following ARF induction. The average area of autophagosomes, calculated with ImageJ software, is indicated below each panel. The data depicted are representative of three independent experiments, in multiple independent clones of each mutant. Scale bar: 500 nm. (D) Western blot analysis of U2OS-ARF cells containing wild type ARF (wt) or the valine-to-alanine mutant at codon 113 (V113A), untreated or treated with doxycycline (doxy, 0.1 μg/ml) for 24 h. Degradation of SQSTM1 and accumulation of LC3-II are indicators of autophagy induction. Actin (ACTB) is included as a loading control. The data depicted are representative of three independent experiments.
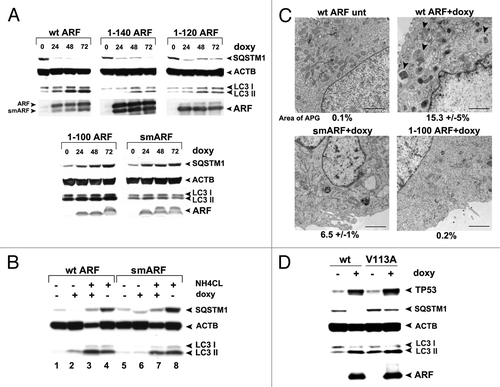
Figure 4. smARF induces morphological indicators of mitophagy. (A) Electron microscopy of U2OS-smARF cells untreated or treated with doxycycline for 48 h (0.1 µg/mL). The arrowheads indicate swollen mitochondria evident after smARF induction. The data depicted are representative of several independent experiments, and two different subclones of U2OS-smARF cells. Scale bar: 500 nm. (B) U2OS-smARF cells were treated with either doxycycline (doxy, 0.1 µg/mL) or a positive control (the mitochondrial uncoupler carbonyl cyanide m-chlorophenylhydrazone, CCCP, 10 μM) for 48 h or left untreated (unt). The mitochondrial transmembrane potential was measured using the flow cytometry-based MitoPotential assay from Millipore (Guava EasyCyte). Note that induction of smARF increases the percentage of cells with depolarized mitochondria. The graph depicts data from three independent experiments, with standard error. (C) U2OS-ARF cells were transiently transfected with GFP-LC3 plasmid for 24 h, treated with doxycycline for 48 h (0.1 µg/mL) and examined by confocal microscopy. The chart shows the percentage of cells containing more than four GFP-LC3 vacuoles compared with untreated controls. The results are the average plus standard error of three independent experiments in which at least 200 cells were counted for each sample. (D) Immunofluorescence analysis of U2OS-smARF cells transiently transfected with GFP-LC3 plasmid for 24 h, then untreated or treated with doxycycline for 48 h (0.1 µg/mL) and stained with MitoTracker Red. The bottom panels depict the colocalization of GFP-LC3 vacuoles with MitoTracker stain following smARF induction. (E) U2OS cells with inducible smARF were left untreated or incubated with 0.1 µg/mL doxycycline for 48 h, and mitochondrial mass was assessed following incubation with MitoTracker Green (MTG). The data depicted are representative of three independent experiments. (F) U2OS cells with inducible smARF were left untreated or incubated with 0.1 µg/mL doxycycline for 24 h, and then analyzed by immunofluorescence for TOMM20. The data depicted are representative of three independent experiments.
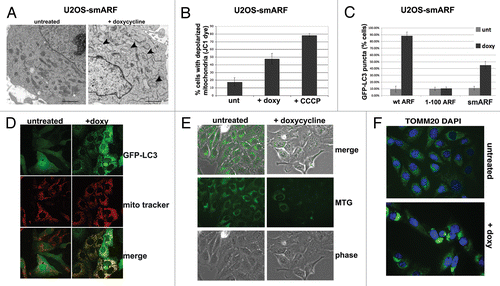
Figure 5. A conserved region in the C terminus of ARF is required for autophagy in both human and mouse. (A) Schematic representation of deletion mutants of human ARF p14ARF (ARF) that were cloned into the U2OS/Tet-On-inducible cell line (U2OS-TREx). To keep the numbering of human ARF consistent with other studies, the extended transcript is denoted −41/+132. (B) Protein levels of ARF deletion mutants, and level of TP53 induced by each mutant, in inducible cells following 24 h in doxycycline (doxy, 0.1 µg/mL). Note that the monoclonal antibody that is used to detect ARF in the left two samples (−41/+132 and 1–132) does not recognize the −41/+99 and −41/+59 deletion mutants; therefore, for the right two samples (−41/+99 and −41/+59), a different antibody, which maps to the N-terminus of ARF, was used (Abcam, Ab14930). (C) Western blot analysis of SQSTM1 in U2OS cells following induction of full-length ARF (−41/+132), the −41/+59 mutant, and the 1 to 132 protein. Actin (ACTB) is indicated as a loading control. Note that the −41/+59 mutant lacks the epitope detected by the Sigma ARF antibody. (D) Electron microscopy of U2OS-ARF cells untreated or treated with doxycycline (0.1 µg/mL) for 48 h. The average and standard deviation of the area of autophagosomes (APG), calculated with ImageJ software, is indicated on the bottom. The results were consistent in three independent experiments, using two clones for each mutant. Scale bar: 500 nm. (E) U2OS-ARF cells were transiently transfected with GFP-LC3 plasmid for 24 h and untreated or treated with doxycycline for 48 h (0.1 µg/mL), and then analyzed by confocal microscopy. Cytoplasmic GFP-LC3 vesicles were detectable in full-length (−41/+132) ARF cells and for the 1 to 132 version, but not in cells expressing the −41/+99 deletion mutant. (F) The percentage of cells with four or more GFP-LC3 vacuoles in (E) was quantified in three independent experiments; error bars mark standard error.
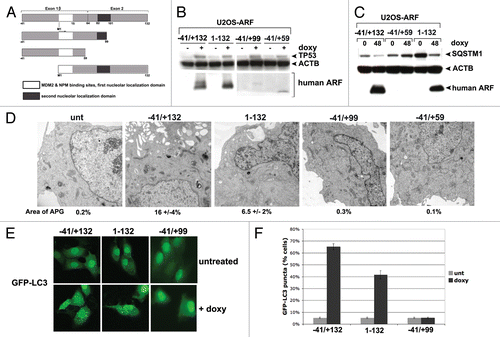
Figure 6. Tumor-derived mutants of ARF are impaired for autophagy. (A) Western blot analysis of U2OS-ARF cells containing wild-type ARF (−41/+132) or the mutants indicated, untreated or treated with doxycycline (doxy, 0.1 μg/ml) for 24 h. Stabilization of TP53 and accumulation of LC3-II are indicated. Actin (ACTB) is included as a loading control. The data depicted are representative of three independent experiments, for multiple clones of each mutant. (B) U2OS-ARF cells containing the mutants indicated were transiently transfected with GFP-LC3 plasmid for 24 h and untreated or treated with doxycycline for 24 h (0.1 µg/mL), and then analyzed by confocal microscopy for GFP-LC3 vesicles. Cytoplasmic GFP-LC3 vesicles were detectable in the P94L mutant, but not in the R98L, L104I and R115G mutants. (C) Electron microscopy of autophagosomes in U2OS-ARF cells containing the ARF mutants indicated, or parental cells (U2OS-TRex), untreated or treated with doxycycline (0.1 µg/mL) for 48 h. The results depicted were consistent in three independent experiments, using two clones for each mutant. Scale bar: 500 nm.
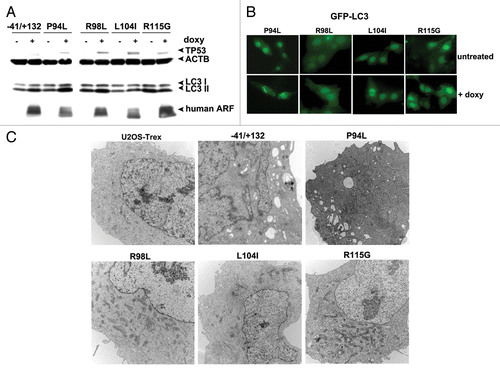
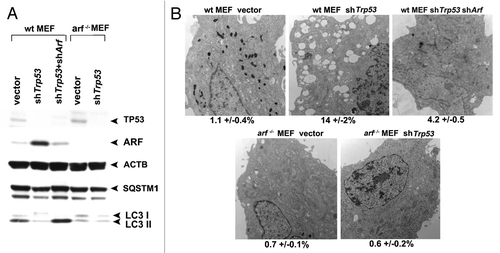
Figure 7. Autophagy induced by Trp53 silencing requires ARF. (A) Primary mouse embryo fibroblasts (wt MEFs) and MEFs from the Arf knockout mouse (arf−/− MEFs) were infected with parental MLP retrovirus (vector), retroviral short hairpin to Trp53 (shTrp53), or retroviral short hairpin to Arf (shArf). Whole cell lysates following infection and selection were subjected to western blot analysis using the antibodies indicated. The level of autophagy induced is assessed by the degradation of SQSTM1; the lower molecular weight SQSTM1 band is thought to be a degradation product. Note that silencing of Trp53 in arf−/− MEFs, and silencing of both Trp53 and Arf in wt MEFs, does not induce appreciable autophagy. The data presented are representative of three independent experiments, in two independent batches of MEFs from each genotype. (B) Electron microscopy of wild-type and arf−/− MEFs following infection with the retroviruses indicated. The average area of autophagosomes calculated with ImageJ software, with standard error, is indicated on the bottom of each panel. Scale bar: 500 nm.