Figures & data
Figure 1. A23187 (A23) and thapsigargin (TG) block autophagic degradation and sequestration activity under both nutrient-rich and amino acid starvation conditions. LNCaP cells (A) or U2OS cells (B) were radiolabeled with [14C]-valine for 2 to 3 d and chased for 18 h. Subsequently, cells were incubated in complete medium containing 0.2% DMSO (vehicle control; Ctrl), A23 (2 µM) or TG (100 nM) for either 6 h or 16 h as indicated, and the degradation of long-lived proteins was determined as described in Materials and Methods. The columns represent mean degradation rates ± standard error of the mean (SEM) from 3 independent experiments. **P < 0.01 by the Student t-test. (C and D) Degradation of long-lived proteins was measured in LNCaP (C) cells during a 6 h incubation or in U2OS (D) cells during a 16 h incubation with DMSO (Ctrl, 0.1%), A23 (2 µM) or TG (100 nM) alone or in combination with DMSO (Ctrl, 0.1%), BafA1 [Baf, 100 nM in (C); 20 nM in (D)], chloroquine [(CQ, 100 µM in (C); 50 µM in (D)] or MG132 [(MG, 5 µM in (C); 1 µM in (D)] as indicated. Columns represent mean relative degradation rates ± SEM from 3 independent experiments, arbitrarily setting the mean degradation rate in the DMSO/DMSO control condition to 1. **P < 0.01 by the Student t-test. (E and F) LNCaP cells (E) or U2OS cells (F) were radiolabeled with [14C]-valine and chased as in (A). Subsequently, cells were incubated for 6 h in either complete medium (CM) containing DMSO (Ctrl, 0.2%) or in medium devoid of serum and amino acids (EBSS) in the presence of DMSO (Ctrl, 0.2%), A23 (0.75 µM) or TG (100 nM), and the degradation of long-lived proteins during this period was measured. Columns represent mean protein degradation rates ± SEM from 6 (E) or 7 (F) independent experiments. **P < 0.01 by the Student t-test. Note that a lower concentration of A23 (0.75 µM) than that used in (A–D) was sufficient to block the degradation of long-lived proteins induced by amino acid starvation. (G) LNCaP cells were incubated for 3 h or 16 h in complete medium containing DMSO (Ctrl, 0.1%), A23 (2 µM), TG (100 nM) or 3-MA (10 mM) with the additional presence of DMSO (Ctrl, 0.1%) or BafA1 (100 nM for 3 h or 20 nM for 16 h) as indicated. Subsequently, cells were harvested and the amount of sequestered lactate dehydrogenase (LDH) was measured as detailed in Materials and Methods. Mean values from 4 (3 h) or 3 (16 h) independent experiments are shown, with error bars representing SEM *P < 0.05, **P < 0.01 by the Student t-test. (H and I) LNCaP cells (H) or U2OS cells (I) were incubated for 2 h in complete medium containing DMSO (Ctrl, 0.2%) or in medium devoid of serum and amino acids (EBSS) containing DMSO (Ctrl, 0.1%), BafA1 (100 nM), A23 (0.75 µM), TG (100 nM), or 3-MA (10 mM) as indicated. Subsequently, cells were harvested and the amount of sequestered LDH was measured as detailed in Materials and Methods. Mean relative values from five independent experiments are shown, with error bars representing SEM *P < 0.05, **P < 0.01 by the Student t-test.
![Figure 1. A23187 (A23) and thapsigargin (TG) block autophagic degradation and sequestration activity under both nutrient-rich and amino acid starvation conditions. LNCaP cells (A) or U2OS cells (B) were radiolabeled with [14C]-valine for 2 to 3 d and chased for 18 h. Subsequently, cells were incubated in complete medium containing 0.2% DMSO (vehicle control; Ctrl), A23 (2 µM) or TG (100 nM) for either 6 h or 16 h as indicated, and the degradation of long-lived proteins was determined as described in Materials and Methods. The columns represent mean degradation rates ± standard error of the mean (SEM) from 3 independent experiments. **P < 0.01 by the Student t-test. (C and D) Degradation of long-lived proteins was measured in LNCaP (C) cells during a 6 h incubation or in U2OS (D) cells during a 16 h incubation with DMSO (Ctrl, 0.1%), A23 (2 µM) or TG (100 nM) alone or in combination with DMSO (Ctrl, 0.1%), BafA1 [Baf, 100 nM in (C); 20 nM in (D)], chloroquine [(CQ, 100 µM in (C); 50 µM in (D)] or MG132 [(MG, 5 µM in (C); 1 µM in (D)] as indicated. Columns represent mean relative degradation rates ± SEM from 3 independent experiments, arbitrarily setting the mean degradation rate in the DMSO/DMSO control condition to 1. **P < 0.01 by the Student t-test. (E and F) LNCaP cells (E) or U2OS cells (F) were radiolabeled with [14C]-valine and chased as in (A). Subsequently, cells were incubated for 6 h in either complete medium (CM) containing DMSO (Ctrl, 0.2%) or in medium devoid of serum and amino acids (EBSS) in the presence of DMSO (Ctrl, 0.2%), A23 (0.75 µM) or TG (100 nM), and the degradation of long-lived proteins during this period was measured. Columns represent mean protein degradation rates ± SEM from 6 (E) or 7 (F) independent experiments. **P < 0.01 by the Student t-test. Note that a lower concentration of A23 (0.75 µM) than that used in (A–D) was sufficient to block the degradation of long-lived proteins induced by amino acid starvation. (G) LNCaP cells were incubated for 3 h or 16 h in complete medium containing DMSO (Ctrl, 0.1%), A23 (2 µM), TG (100 nM) or 3-MA (10 mM) with the additional presence of DMSO (Ctrl, 0.1%) or BafA1 (100 nM for 3 h or 20 nM for 16 h) as indicated. Subsequently, cells were harvested and the amount of sequestered lactate dehydrogenase (LDH) was measured as detailed in Materials and Methods. Mean values from 4 (3 h) or 3 (16 h) independent experiments are shown, with error bars representing SEM *P < 0.05, **P < 0.01 by the Student t-test. (H and I) LNCaP cells (H) or U2OS cells (I) were incubated for 2 h in complete medium containing DMSO (Ctrl, 0.2%) or in medium devoid of serum and amino acids (EBSS) containing DMSO (Ctrl, 0.1%), BafA1 (100 nM), A23 (0.75 µM), TG (100 nM), or 3-MA (10 mM) as indicated. Subsequently, cells were harvested and the amount of sequestered LDH was measured as detailed in Materials and Methods. Mean relative values from five independent experiments are shown, with error bars representing SEM *P < 0.05, **P < 0.01 by the Student t-test.](/cms/asset/fbf85389-201a-4b8e-bbc9-ae6c6ca9fb52/kaup_a_10925900_f0001.gif)
Figure 2. A23187 (A23) and TG rapidly and persistently induce ER stress in the absence of caspase activation. LNCaP cells (A–F) or U2OS cells (G–L) were incubated with DMSO (0.2%; vehicle control), A23 (2 µM) or TG (100 nM) for various lengths of time as indicated, or BafA1 (100 nM) for 24 h as a positive control for ER stress induction and caspase activation. Subsequently, cell lysates were prepared and subjected to western blot analyses of EIF2AK3, phopsho-EIF2S1 (p-EIF2S1), ATF4, XBP1s, DDIT3, HSPA5, PARP, cleaved CASP3 (cl-CASP3) or EIF2S1 (loading control) as indicated. Representative immunoblots from 4 independent experiments are shown in (A and G), and mean relative protein levels normalized to the loading control are shown in (B–F) and (H–L). The mean values obtained in DMSO control-treated cells were arbitrarily set to 1, except for in (K) where the mean value for the 2 h treatment condition with TG was arbitrarily set to 1, as this was the first time point at which a persistent quantifiable signal appeared. Error bars represent SEM-values. The observed shift toward the more slowly migrating bands that are recognized by the EIF2AK3 antibody is indicative of EIF2AK3 phosphorylation. This was confirmed when using a specific EIF2AK3 inhibitor (see , middle panels). The caspase-specific 89-kDa cleavage product of PARP is indicated by “cl-PARP”.
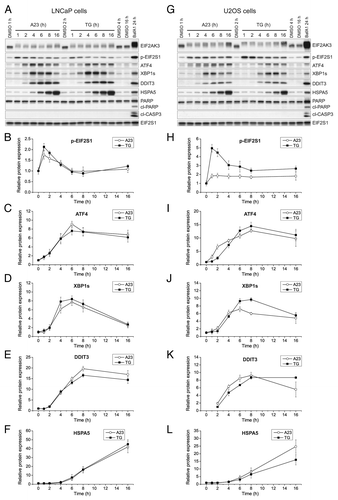
Figure 3. A23187 (A23)- and TG-mediated repression of autophagy is independent of ER stress. LNCaP cells (A) or U2OS cells (B) were transfected with a nontargeting control siRNA (siCtrl) or siRNAs targeting ATF4 (siATF4), ERN1 (siERN1), or ATF6 (siATF6). Subsequently, cells were treated for 6 h with DMSO control (Ctrl, 0.2%), A23 (0.75 µM), or TG (100 nM) under amino acid starvation conditions, and long-lived protein degradation rates were determined as described in Materials and Methods. Mean protein degradation rates ± SEM from 3 independent experiments are shown. (C and D) Confirmation of knockdown efficiencies of the siRNAs used in (A and B). LNCaP cells (C) or U2OS cells (D) were transfected with siRNAs as in (A and B) and treated for 6 h with DMSO (Ctrl, 0.2%) or TG (100 nM) under amino acid starvation conditions. Subsequently, RNA was extracted and subjected to real-time RT-PCR analyses of relative mRNA levels of ATF4, ERN1, and ATF6 as indicated. Mean relative mRNA levels ± SEM from 3 independent experiments are shown. The average value of the DMSO control conditions with the control siRNA was arbitrarily set to 1. The mean knockdown efficiency varied between 83% and 92%. (E and F) LNCaP cells (E) or U2OS cells (F) were pretreated for 1 h with DMSO (Ctrl, 0.2%), the EIF2AK3 inhibitor GSK2606414 (EIF2AK3i, 100 nM) + 0.1% DMSO, the ERN1-inhibitor MKC8866 (ERN1i, 10 µM) + 0.1% DMSO, or a combination of the two inhibitors (100 nM EIF2AK3i + 10 µM ERN1i). Thereafter, the cells were washed in EBSS medium and starved for 2 h in EBSS medium containing BafA1 (100 nM) and either DMSO (Ctrl, 0.1%), A23 (0.75 µM) or TG (100 nM). Moreover, to maintain suppression of EIF2AK3 and ERN1 activity, EIF2AK3i, ERN1i, and DMSO were re-added at the same concentrations and combinations as during the pretreatment. Subsequently, cells were harvested and assessed for LDH sequestration (top panels) as detailed in Materials and Methods. Mean relative LDH sequestration values ± SEM from 3 independent experiments are shown. The mean LDH sequestration rate in cells that were starved for amino acids in the absence of BafA1 was arbitrarily set to 1. Second (middle panels), aliquots of the total disruptates that were generated in the LDH sequestration assay were used for western blot analyses of EIF2AK3, XBP1s, and GAPDH (loading control) as detailed in Materials and Methods. The observed shift toward the more slowly migrating bands that are recognized by the EIF2AK3 antibody is indicative of EIF2AK3 phosphorylation. Third (bottom panels), aliquots of the harvested cells were subjected to real-time RT-PCR analysis of XBP1s mRNA levels as described in Materials and Methods. Mean relative values ± SEM from the 3 independent experiments are shown. (G and H) [14C]-valine radiolabelled LNCaP cells (G) or U2OS cells (H) were chased and then treated as in (E and F), but for 6 h and without BafA1 addition. Degradation rates of long-lived proteins were determined as detailed in Materials and Methods. The columns represent mean degradation rates ± SEM from 3 (G) or 4 (H) independent experiments.
![Figure 3. A23187 (A23)- and TG-mediated repression of autophagy is independent of ER stress. LNCaP cells (A) or U2OS cells (B) were transfected with a nontargeting control siRNA (siCtrl) or siRNAs targeting ATF4 (siATF4), ERN1 (siERN1), or ATF6 (siATF6). Subsequently, cells were treated for 6 h with DMSO control (Ctrl, 0.2%), A23 (0.75 µM), or TG (100 nM) under amino acid starvation conditions, and long-lived protein degradation rates were determined as described in Materials and Methods. Mean protein degradation rates ± SEM from 3 independent experiments are shown. (C and D) Confirmation of knockdown efficiencies of the siRNAs used in (A and B). LNCaP cells (C) or U2OS cells (D) were transfected with siRNAs as in (A and B) and treated for 6 h with DMSO (Ctrl, 0.2%) or TG (100 nM) under amino acid starvation conditions. Subsequently, RNA was extracted and subjected to real-time RT-PCR analyses of relative mRNA levels of ATF4, ERN1, and ATF6 as indicated. Mean relative mRNA levels ± SEM from 3 independent experiments are shown. The average value of the DMSO control conditions with the control siRNA was arbitrarily set to 1. The mean knockdown efficiency varied between 83% and 92%. (E and F) LNCaP cells (E) or U2OS cells (F) were pretreated for 1 h with DMSO (Ctrl, 0.2%), the EIF2AK3 inhibitor GSK2606414 (EIF2AK3i, 100 nM) + 0.1% DMSO, the ERN1-inhibitor MKC8866 (ERN1i, 10 µM) + 0.1% DMSO, or a combination of the two inhibitors (100 nM EIF2AK3i + 10 µM ERN1i). Thereafter, the cells were washed in EBSS medium and starved for 2 h in EBSS medium containing BafA1 (100 nM) and either DMSO (Ctrl, 0.1%), A23 (0.75 µM) or TG (100 nM). Moreover, to maintain suppression of EIF2AK3 and ERN1 activity, EIF2AK3i, ERN1i, and DMSO were re-added at the same concentrations and combinations as during the pretreatment. Subsequently, cells were harvested and assessed for LDH sequestration (top panels) as detailed in Materials and Methods. Mean relative LDH sequestration values ± SEM from 3 independent experiments are shown. The mean LDH sequestration rate in cells that were starved for amino acids in the absence of BafA1 was arbitrarily set to 1. Second (middle panels), aliquots of the total disruptates that were generated in the LDH sequestration assay were used for western blot analyses of EIF2AK3, XBP1s, and GAPDH (loading control) as detailed in Materials and Methods. The observed shift toward the more slowly migrating bands that are recognized by the EIF2AK3 antibody is indicative of EIF2AK3 phosphorylation. Third (bottom panels), aliquots of the harvested cells were subjected to real-time RT-PCR analysis of XBP1s mRNA levels as described in Materials and Methods. Mean relative values ± SEM from the 3 independent experiments are shown. (G and H) [14C]-valine radiolabelled LNCaP cells (G) or U2OS cells (H) were chased and then treated as in (E and F), but for 6 h and without BafA1 addition. Degradation rates of long-lived proteins were determined as detailed in Materials and Methods. The columns represent mean degradation rates ± SEM from 3 (G) or 4 (H) independent experiments.](/cms/asset/48bbf023-a00a-4e7d-948e-0eeb7910b888/kaup_a_10925900_f0003.gif)
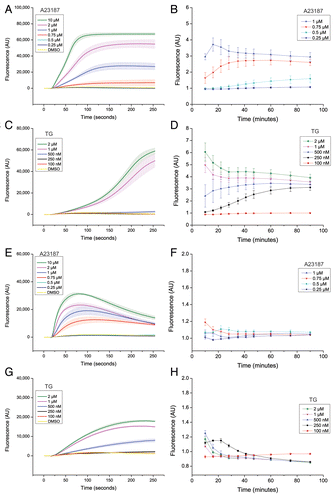
Figure 4. Suppression of autophagy does not require bulk or sustained elevation of cytosolic calcium levels. LNCaP cells (A–D) or U2OS cells (E–H), loaded with the calcium-sensitive dye “FLIPR calcium V”, were treated simultaneously at the 16 s time point with either DMSO (0.2%; vehicle control) or various concentrations of A23187 or TG as indicated. Changes in cytosolic calcium ion levels were followed over a period of 90 min as detailed in Materials and Methods, first by using the FLIPR384 instrument for imaging at 1 s intervals for the first 4 min after treatment (A, C, E, and G), and second by using a Tecan Infinite F200 Pro plate reader for fluorescence measurements at various time intervals from 10 to 90 min (B, D, F, and H). Each panel shows mean values ± SEM at each time point from the 3 independent experiments. Each experiment was performed in six replicate wells. The graphs in (A, C, E, and G) shows mean values ± SEM of the raw fluorescence values obtained in DMSO- A23187- or TG-treated cells at each time point. In (B, D, F, and H), the mean value obtained in DMSO control-treated cells was arbitrarily set to 1 at each time point. AU = arbitrary units. Of note, unlike in complete culture medium, the two highest concentrations of A23187 (2 and 10 µM) showed signs of toxic effects under amino acid and serum starvation conditions (in EBSS or in the HBSS-HEPES buffer solution used in the calcium measurement assay). Long-term calcium measurements for these two concentrations of A23187 are therefore not shown in (B and F), and were also not used to assess effects of A23187 on autophagy under conditions of amino acid starvation. Also note that several of the lines representing measurements of the DMSO control and concentrations of A23187 and TG which give no discernible increase in fluorescence compared with the DMSO control condition (i.e., no detectable increase in cytosolic calcium levels) often overlap.