Figures & data
Figure 1. Autophagy induction by Aβ42 is impaired by PRNP deficiency. (A) HpL3-4(PrP) and HpL3-4(E) cells were transfected with mCherry-GFP-LC3 for 24 h and then incubated in serum-free medium (Starved) or treated with 0.5 μM Aβ42 for 12 h. The cells were stained with Hoechst dye and observed under a confocal microscope. Scale bars: 10 μm. (B) The number of cells with mCherry-LC3 and/or GFP-LC3 puncta in confocal images, including (A), was quantified (mean values ± S.D., n = 12; *P < 0.01, **P < 0.005 vs. control cells; ns, not-significant). (C) HpL3-4(PrP) and HpL3-4(E) cells were incubated with 0.5 μM Aβ42 for 12 h and then subjected to electron microscopic analysis. Asterisks indicate autophagic vacuoles (AVs). AV and mitochondria (M) are also shown in enlarged view. (D) The numbers of cytoplasmic AVs were calculated (mean values ± S.D., n = 20; **P < 0.005 vs. control cells). (E) HpL3-4(PrP) and HpL3-4(E) cells were incubated in serum-free medium (Starved) or treated with 0.5 μM Aβ42 for 12 h in the absence or presence of 20 nM bafilomycin A1 (BafA1) for 3 h before harvesting, and the cell lysates were analyzed by western blotting using the indicated antibodies. (F) Three-month-old WT and prnp-KO mice were injected i.c.v. with PBS or 2 μg Aβ42. After 12 and 48 h, hippocampal lysates were prepared and subjected to western blot. The signals on the blot were quantified using densitometric analysis and the relative ratios of LC3-II to ACTB are shown at the bottom of the blot.
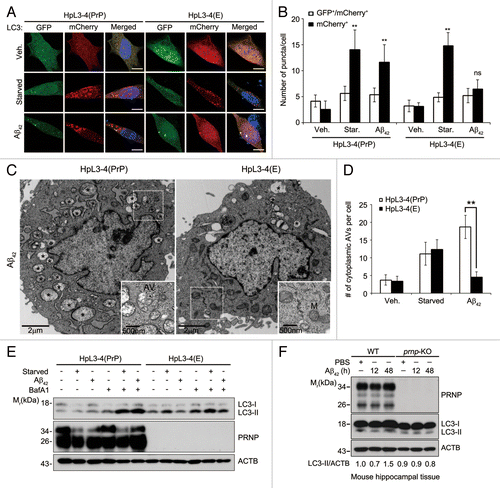
Figure 2. Aβ42 enhances PIK3C3 complex activity via PRNP. (A) SH-SY5Y cells were transfected with FYVE-RFP and either pSuper-neo or PRNP shRNA for 48 h and then left untreated (Veh.) or incubated with either serum-free medium for 12 h (Starved) or 0.5 μM Aβ42 for 12 h. The cells were observed under a confocal microscope (upper) and the number of FYVE-RFP puncta per cell was quantified. The bars represent mean values ± S.D. (n = 30 cells) (lower). The arrowheads indicate FYVE-RFP puncta. Scale bars: 10 μm. (B and C) HpL3-4(PrP) and HpL3-4(E) cells were left untreated (Veh.) or incubated with 5 mM 3MA for 3 h, EBSS for 3 h, or 0.5 μM Aβ42 for 12 h. Cell lysates were subjected to an immunoprecipitation assay using anti-BECN1 antibody. The immunoprecipitates bound to protein G-beads were used for the PtdIns3K assay (B) and the immunoprecipitated BECN1 was detected with western blotting using anti-BECN1 antibody (C). The signal of BECN1 on the blot was quantified by densitometric analysis and used to normalize the PtdIns3K activity. The asterisk indicates nonspecific signal and bars represent mean values ± S.E. (n = 3).
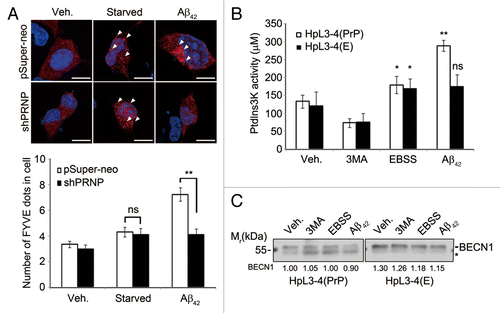
Figure 3. PRNP interacts with BECN1. (A and B) HEK293T cells were cotransfected with PRNP-GFP and BECN1-HA (A) or PRNP-HA and BECN1 (B) for 24 h, after which the cell lysates were subjected to an immunoprecipitation (IP) assay using an anti-HA antibody (bottom). The immunoprecipitates were then analyzed with western blotting using anti-GFP (A) and anti-BECN1 (B) antibodies (top). Whole cell lysates (WCLs) were examined with western blotting as a reference. (C) Tissue lysates prepared from the cortex of 3-mo-old C57BL/6 mice were analyzed with an immunoprecipitation (IP) assay using preimmune serum of rat (control) or anti-BECN1 antibody (#1: From Novus; #2: This study). The immunoprecipitates were examined with immunoblotting using anti-PRNP and anti-BECN1 antibodies. The arrows and asterisks indicate PRNP and non-specific signals, respectively. (D) BECN1 (10 μg) purified from E. coli was separated by SDS-PAGE and transferred to a PVDF membrane. After an overnight incubation with 3% BSA or mouse whole brain lysates, the membrane was subjected to western blot analysis using the indicated antibodies. Bovine serum albumin (BSA; 10 μg) was used as a control. (E) SH-SY5Y cell lysates were prepared and analyzed with an immunoprecipitation (IP) assay using anti-rabbit serum (control) and anti-CAV1 antibody. (F) SH-SY5Y cells were cotransfected with CAV1-HA, PRNP and BECN1for 24 h and the cell lysates were then subjected to a co-IP assay using anti-HA antibodies. The immunoprecipitates were analyzed with western blotting using the indicated antibodies. (G) SH-SY5Y cells were cotransfected with PRNP, BECN1-HA and CAV1 shRNA (shCAV1), as indicated, for 48 h and the cell lysates were then subjected to an immunoprecipitation assay (IP) using anti-HA antibody. The immunoprecipitates were analyzed with western blotting using the indicated antibodies. WCL, whole cell lysate.
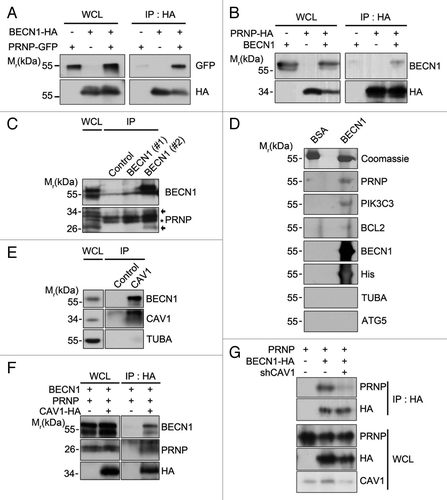
Figure 4. Enhanced localization of BECN1 in lipid rafts by Aβ42 in the presence of PRNP. (A) HpL3-4(PrP) and HpL3-4(E) cells were left untreated (Veh.) or incubated with 0.5 μM Aβ42 for 12 h. Cell homogenates were subjected to sucrose gradient ultracentrifugation. Equal volumes of the fractions were subjected to western blot analysis using the indicated antibodies. (B) The intensity of BECN1 on the western blot images in (A) was quantified (mean values ± S.E., n = 3). (C) HpL3-4(PrP) and HpL3-4(E) cells were transfected with BECN1-GFP (green) for 24 h. After incubation with Alexa Fluor-594-conjugated cholera toxin subunit B (red) for 15 min at 4 °C, the cells were further incubated for 30 min at 37 °C for uptake. Cell nuclei were stained with Hoechst dye (blue) and observed under a confocal microscope. (D and E) HpL3-4(PrP) and HpL3-4(E) cells were left untreated or incubated with 0.5 μM Aβ42 for 12 h, and cell extracts were subjected to sucrose gradient ultracentrifugation as in (A). DRM fractions (fractions #4 to 6) and non-DRM fractions (fractions #11 to 12) were then pooled and subjected to an immunoprecipitation assay using an anti-BECN1 antibody. The immunoprecipitates were assayed for PtdIns3K activity and bars represent mean values ± S.E. (n = 3) (D) or proved with western blotting using anti-BECN1 antibody (E).
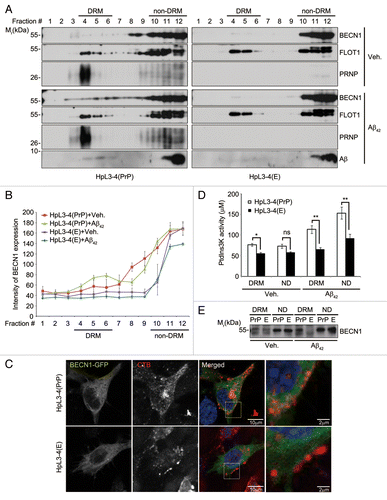
Figure 5. BCL2-binding domain of BECN1 is required for its interaction with PRNP and lipid rafts localization. (A) Schematic diagram of BECN1 domains and its deletion mutants. The binding ability to PRNP and the PtdIns3K activity of BECN1 and its deletions are summarized. (B) HEK293T cells were cotransfected with PRNP and either BECN1-flag (FL), BECN1−BDΔ-flag (BDΔ), BECN1−CCDΔ-flag (CCDΔ), or BECN1−ECDΔ-flag (ECDΔ) for 24 h. Cell lysates were analyzed with immunoprecipitation (IP) using anti-flag beads and immunoblotting using anti-PRNP and anti-flag antibodies. (C) SH-SY5Y cells were transfected with BECN1 or its mutant for 24 h and cell homogenates were subjected to sucrose gradient ultracentrifugation and western blotting as described in . WCL, whole cell lysate.
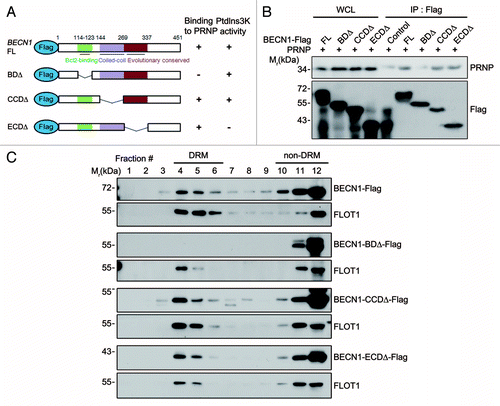
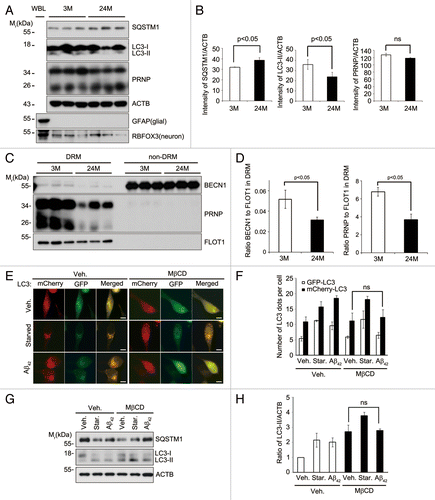
Figure 6. Lipid raft-localized BECN1 and PRNP decrease in aged hippocampus, and disruption of lipid rafts by MβCD impairs Aβ42-induced autophagy. (A) Hippocampal neurons were purified by discontinuous iodixanol gradient assay from 3- and 24-mo-old WT C57BL/6 mice and their lysates and whole brain lysates (WBL) were subjected to western blot analysis using the indicated antibodies. (B) The signals on the blots were quantified by densitometric analysis using the ScienceLab software. The bars represent mean values ± S.E. (n = 3). (C) DRM (fractions #4 to 5) and non-DRM (fractions #11 to 12) fractions of each mouse were pooled and then subjected to western blot analysis. (D) The signals on the blot shown in (C) were quantified by densitometric analysis. The bars represent the relative ratios of BECN1 in the DRM to the non-DRM fractions with mean values ± S.D. (n = 3). (E) SH-SY5Y cells were transiently transfected with mCherry-GFP-LC3 for 24 h and left untreated (Veh.) or treated with 1 mg/mL MβCD for 12 h. The cells were then incubated with serum-free DMEM (Starved) or 1 μM Aβ42 for 12 h, and then observed under a fluorescence microscope. Scale bars: 10 μm. (F) The number of mCherry-LC3- and GFP-LC3-positive puncta per cell was counted and represented as mean values ± S.D. (n = 12). (G) SH-SY5Y cells were pretreated with PBS (Veh.) or 1 mg/mL MβCD for 12 h and then incubated with serum-free DMEM (Starved) or 1 μM Aβ42 for 3 h. Cell extracts were analyzed with western blotting using anti-SQSTM1, anti-LC3, and anti-ACTB antibodies. (H) The signals on the blots were quantified by densitometric analysis. The bars represent the relative ratios of the LC3-II signal to ACTB with mean values ± S.D. (n = 3).