Figures & data
Figure 1. Atg32 undergoes starvation-dependent processing. (A) Cells expressing TAP tagged Atg32 under the control of the GAL1 promoter (KWY100) were cultured in YPG to mid-log phase and shifted to SD-N for the indicated times. TAP-Atg32 was monitored by immunoblotting with an antibody that recognizes PA. The arrowhead here and in the following panels indicates the processed band. (B) Cells expressing GAL1 promoter-driven TAP-Atg32 in wild-type (WT; KWY100), atg1∆ (KWY101), and atg11∆ (KWY104) backgrounds were cultured in YPG to mid-log phase and shifted to SD-N for the indicated times. (C) Cells expressing endogenous promoter-driven TAP-Atg32 in a WT background (KWY139) were cultured in YPL to mid-log phase and shifted to SD-N for the indicated times.
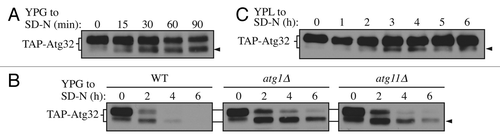
Figure 2. C-terminal tagging of Atg32 interferes with its processing and mitophagy. (A) Strains expressing GAL1 promoter-driven TAP-Atg32 (KWY100) and TAP-Atg32-GFP (KWY110) were cultured as in . Proteins were monitored by immunoblotting with an antibody that detects PA. (B) Wild-type (WT; KWY90), atg32∆ (KWY22), and Atg32-GFP (KWY121) strains expressing mitoPho8∆60 were grown in YPL and shifted to SD-N for the indicated times. Samples were collected and protein extracts were assayed for mitoPho8∆60 activity. The results represent the mean and standard deviation (SD) of three independent experiments. (C) Strains expressing GAL1 promoter-driven N-terminal GFP-tagged Atg32 (KWY111) and C-terminal GFP-tagged Atg32 (KWY121) were cultured in YPGal to mid-log phase and stained with the mitochondrial marker MitoTracker Red. The localization of GFP and MitoTracker Red were visualized by fluorescence microscopy. DIC, differential interference contrast. Scale bar: 5 µm.
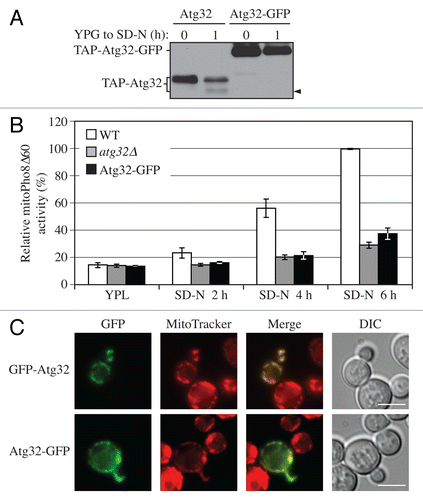
Figure 3. Yme1 mediates Atg32 processing. (A) Cells expressing GAL1 promoter-driven TAP-Atg32 in wild-type (WT; KWY100) and yme1∆ (KWY118) strains were cultured as in . TAP-Atg32 was monitored by immunoblotting with an antibody that binds to PA. Pgk1 was monitored by immunoblotting with anti-Pgk1 antibody as a loading control. (B) Cells expressing GAL1 promoter-driven TAP-Atg32 and Yme1-GFP (KWY134) or Yme1E541Q-GFP (KWY141) were cultured as in . TAP-Atg32 was monitored by immunoblotting with anti-Atg32 antiserum. Yme1-GFP was monitored by immunobloting with anti-YFP antibody. (C) A strain expressing Yme1-GFP (KWY133) was transformed with a plasmid expressing protein A only, or PA-Atg32 under the control of the CUP1 promoter, and cultured in SML medium to mid-log phase or starved in SD-N for 0.5 h. IgG-Sepharose was used to precipitate PA-Atg32 from cell lysates. The bottom two panels show the immunoblot of total cell lysates (input) and the upper two panels show the IgG precipitates (IP), which were probed with anti-YFP antibody and an antibody that recognizes PA. (D) Cells expressing endogenous promoter-driven TAP-Atg32 in WT (KWY139) and yme1∆ (KWY140) strains were cultured as in . TAP-Atg32 was monitored by immunoblotting with an antibody that recognizes protein A.
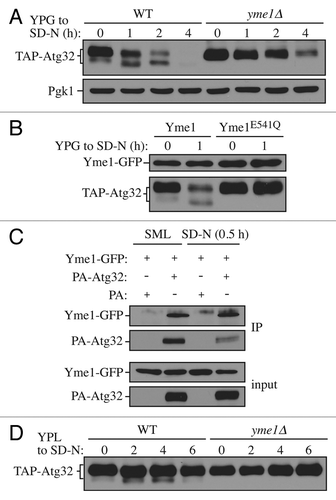
Figure 4. Yme1 is specifically required for mitophagy. (A) Wild-type (WT; KWY20), yme1∆ (KWY136), and atg32∆ (KWY22) strains expressing mitoPho8∆60 were grown in YPL and shifted to SD-N for the indicated times. Samples were collected and protein extracts were assayed for mitoPho8∆60 activity. The results represent the mean and standard deviation (SD) of three independent experiments. (B) Wild-type (KWY20), yme1∆ (KWY136), atg32∆ (KWY22), and Yme1E541Q-GFP (KWY138) strains expressing mitoPho8∆60 were grown in YPL and shifted to SD-N for 6 h. Samples were collected and protein extracts were assayed for mitoPho8∆60 activity. The results represent the mean and standard deviation (SD) of three independent experiments. (C) Wild-type (WLY176), yme1∆ (KWY143), and atg1∆ (WLY192) strains expressing Pho8∆60 were cultured in YPD to mid-log phase and shifted to SD-N for 4 h. Samples were collected and protein extracts were assayed for Pho8∆60 activity. The results represent the mean and standard deviation (SD) of three independent experiments. (D) Wild-type (WLY176), yme1∆ (KWY143), and atg1∆ (WLY192) strains were cultured in YPD medium and analyzed for prApe1 maturation by immunoblotting to monitor the Cvt pathway during vegetative growth. The positions of precursor and mature Ape1 are indicated.
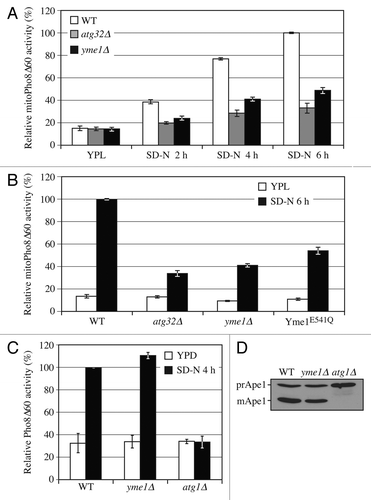
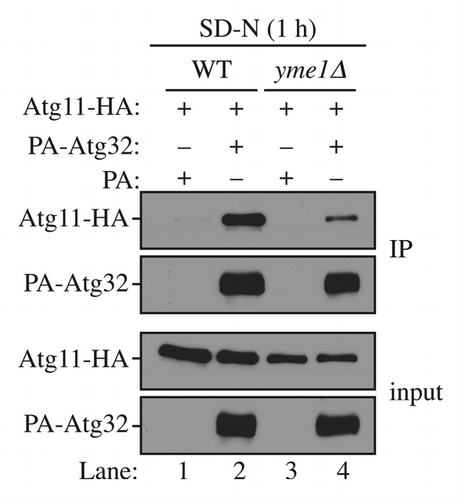
Figure 5. Yme1 regulates the interaction between Atg32 and Atg11. Wild-type (WT; SEY6210) and yme1∆ (KWY142) strains transformed with a plasmid expressing HA-tagged Atg11 together with a plasmid expressing protein A only or PA-Atg32 were cultured in SMD medium to mid-log phase or starved in SD-N for 1 h. IgG-Sepharose was used to precipitate PA-Atg32 from cell lysates. The bottom two panels show the immunoblot of total cell lysates (input) and the upper two panels show the IgG precipitates (IP), which were probed with anti-YFP antibody and an antibody that binds PA. All plasmids are under the control of the CUP1 promoter