Figures & data
Figure 1. Detection of overexpressed Atg13 using rabbit polyclonal antiserum. Yeast cells overexpressing Atg13 were grown in SD and then shifted to SD-N at time 0. Samples were collected at the indicated time points and analyzed by SDS-PAGE and western blot using antiserum to Atg13. This figure is a modification of data previously published in reference Citation8, and is reproduced by permission of the American Society for Biochemistry and Molecular Biology and Elsevier, copyright 2000.
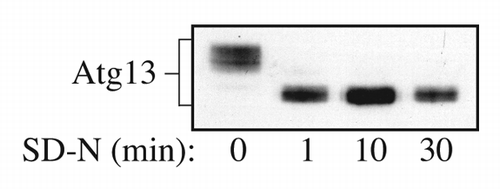
Figure 2. Wild-type yeast cells expressing Atg13 under the endogenous promoter from a multicopy plasmid (YEp351[APG13]) were grown in SD for 8 h and treated with rapamycin for 15 min as indicated. Protein extracts were analyzed by western blot using anti-Atg13 serum.
![Figure 2. Wild-type yeast cells expressing Atg13 under the endogenous promoter from a multicopy plasmid (YEp351[APG13]) were grown in SD for 8 h and treated with rapamycin for 15 min as indicated. Protein extracts were analyzed by western blot using anti-Atg13 serum.](/cms/asset/726a71ef-9cb1-4b9d-8c11-d23892f7a5ef/kaup_a_10927707_f0002.gif)
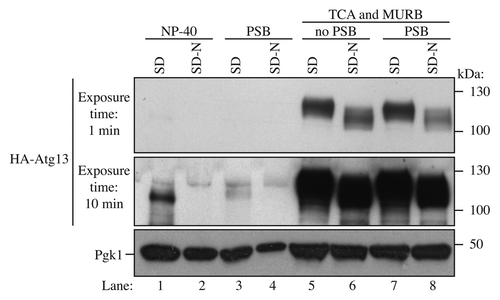
Figure 3. Comparison between different methods to detect Atg13. Wild-type cells (BY4741) were transformed with a vector expressing HA-Atg13 under its own promoter (pHC078). Cultures were grown until mid-log phase in SD and starved for nitrogen 2 h (SD-N). Lanes 1 and 2 correspond to cells that were lysed with NP-40 buffer and disrupted with glass beads; cells were harvested by centrifugation, resuspended in NP-40 lysis buffer (200 µl) containing protease and phosphatase inhibitors, followed by glass bead lysis and centrifugation at 700 × g for 3 min. Proteins were quantified by the BCA assay. The same amount of protein was loaded onto the gel. Lanes 3 and 4 correspond to cells that were disrupted by vortex with glass beads and protein sample buffer (PSB). Lanes 5 to 8 samples were prepared using the TCA-MURB method described in this paper. The protein extracts of lane 7 and 8 were mixed with PSB.