Figures & data
Figure 1. Peroxisomal fission is required for pexophagy. (A) Wild-type yeast cells, transformed with pBFP-SKL (XLY049), were cultured in YTO as described in Materials and Methods to induce peroxisome proliferation, and shifted to SD-N for 2 h. (B) Wild-type (SEY6210), atg1∆ (WHY1), dnm1∆ (KDM1252), fis1∆ (XLY060), vps1∆ (XLY061), and dnm1∆ vps1∆ (XLY062) cells, transformed with pGFP-SKL, were cultured in YTO to induce peroxisome proliferation and shifted to SD-N for 7 h. CellTracker Blue CMAC was used to stain the vacuolar lumen. (C) Quantification of the numbers of peroxisomes in (B). Twelve Z-section images were projected and the number of peroxisomes per cell was determined. Standard deviation was calculated from 3 independent experiments. In (A and C), all of the images are representative pictures from single Z-sections. DIC, differential interference contrast. Scale bar: 2 μm. (D) GFP was tagged at the C terminus of the PEX14 gene on the genome of wild-type (TKYM67), dnm1∆ (KDM1103), fis1∆ (KDM1104), vps1∆ (KDM1105), and dnm1∆ vps1∆ (XLY059) cells. These cells were cultured in YTO to induce peroxisome proliferation and shifted to SD-N for 1 and 2 h. Immunoblotting was done with anti-YFP antibody. The extent of pexophagy was estimated by calculating the amount of free GFP compared with the total amount of intact Pex14-GFP and free GFP.
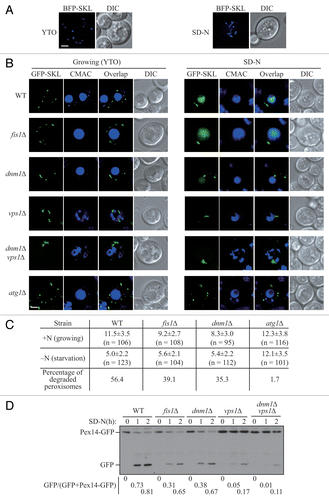
Figure 2. Atg11 recruits the Dnm1 fission complex to peroxisomes. (A) VN-ATG11 PEX14-mCherry (Pex14-mChe; XLY070) cells, transformed with pDnm1-VC, were cultured in YTO to induce peroxisome proliferation and then shifted to SD-N for 1 h. The arrowheads mark colocalizing BiFC and mCherry puncta. (B and D) The plasmids pBFP-SKL and pDnm1-VC were transformed into the VN-ATG11 (KDM1535) cells (B), and pBFP-SKL was transformed into VN-ATG11 VC-FIS1 (KDM1545) cells (D). The arrows mark BiFC puncta colocalizing with MitoTracker Red, arrowheads denote BiFC puncta colocalizing with peroxisomes that are proximal to mitochondria, and asterisks indicate BiFC puncta colocalizing with both peroxisomes and mitochondria. Cells were cultured as described in the Materials and Methods to induce peroxisome proliferation and shifted to SD-N for 1 h. MitoTracker Red was used to stain mitochondria. (C and E) Quantification of the localization of Atg11-Dnm1 interacting puncta (B) and Atg11-Fis1 interacting puncta (D); percentage was calculated based on the number of puncta at a specific location compared with the total number of puncta. In (A, B, and D), all of the images are representative pictures from single Z-sections. DIC, differential interference contrast. Scale bar: 2 μm.
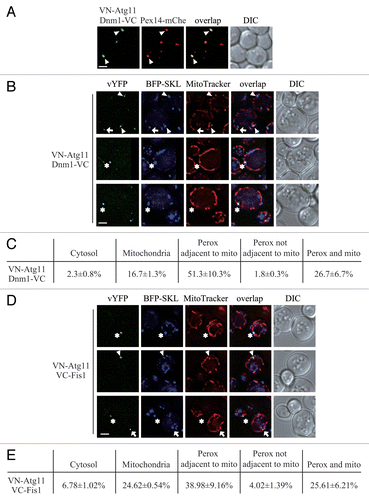
Figure 3. Atg11 recruits Vps1 to peroxisomes that are targeted for degradation. (A) VN-ATG11 PEX14-mCherry (Pex14-mChe; XLY070) cells, transformed with pVps1-VC, were cultured in YTO to induce peroxisome proliferation and subsequently shifted to SD-N for 1 h. (B) The plasmid pCuHA-Atg11 was transformed into atg11∆ (untagged, YTS147), atg11∆ VPS1-PA (KDM1269), atg11∆ DNM1(C30∆)-PA (KDM1249), and atg11∆ VPS1(C30∆)-PA (XLY073) cells. The cells were cultured in YTO to induce peroxisome proliferation and then shifted to SD-N for 2 h. Cell lysates were prepared and incubated with IgG-Sepharose for affinity isolation. The eluted proteins were separated by SDS-PAGE and detected with monoclonal anti-HA antibody and an antibody that binds to PA. (C) The plasmids pBFP-SKL and pVps1-VC were transformed into VN-ATG11 (KDM1535) cells. Cells were cultured in YTO to induce peroxisome proliferation and then shifted to SD-N for 1 h. MitoTracker Red was used to stain mitochondria. (D) Quantification of the localization of Atg11-Vps1 interacting puncta in (C); the percentage was calculated based on the number of puncta at a specific location compared with the total number of puncta. In (A and C) all of the images are representative pictures from single Z-sections. DIC, differential interference contrast. Scale bar: 2 μm.
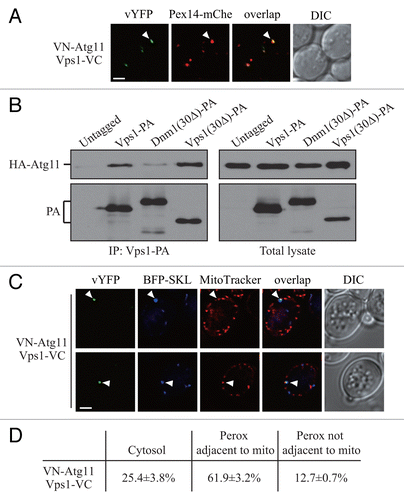
Figure 4. Atg36 interacts with both Dnm1 and Vps1 on the degrading peroxisomes. (A) The plasmid pDnm1-VC or pVps1-VC was transformed into VN-ATG36 (XLY063) cells. These cells together with cells from strains VN-ATG36 VC-FIS1 (XLY064), VN-ATG36 CAF4-VC (XLY065), and VN-ATG36 MDV1-VC (XLY066) were cultured in YTO to induce peroxisome proliferation and then shifted to SD-N for 1 h. (B and D) The plasmid pDnm1-VC was transformed into ATG36-VN pRS405-BFP-SKL (XLY072) cells (B), and pVps1-VC was transformed into ATG36-VN pRS405-BFP-SKL (XLY072) cells (D). Cells were cultured in YTO to induce peroxisome proliferation and then shifted to SD-N for 1 h. MitoTracker Red was used to stain mitochondria. (C and E) Quantification of the localization of Atg36-Dnm1 interacting puncta in (B) and Atg36-Vps1 interacting puncta in (D); the percentage was calculated based on the number of puncta at a specific location compared with the total number of puncta. In (A, B, and D), all of the images are representative pictures from single Z-sections. DIC, differential interference contrast. Scale bar: 2 μm.
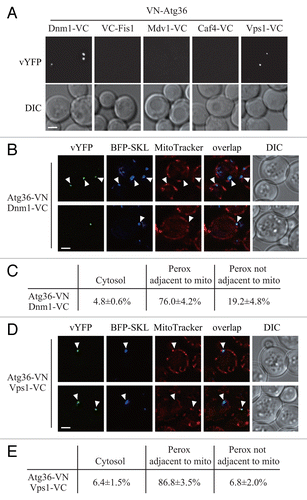
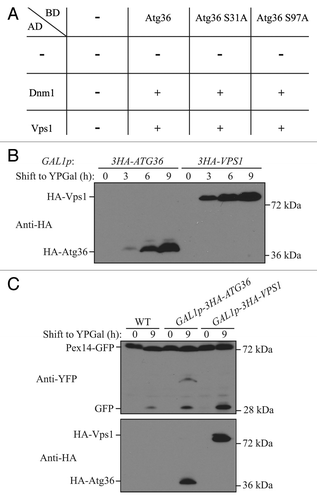
Figure 5. Overexpression of Vps1 induces pexophagy. (A) Yeast 2-hybrid analysis of Atg36 and the indicated mutants interacting with Dnm1 and Vps1. (B and C) GFP was tagged at the C terminus of the PEX14 gene in the genome of wild-type (TKYM67), GAL1p-3HA-ATG36 (XLY088), and GAL1p-3HA-VPS1 (XLY089) cells. These cells were cultured in YTO to induce peroxisome proliferation and shifted to YPG for 3, 6, and 9 h. Immunoblotting was done with anti-HA antibody in (B) to monitor protein expression following galactose induction, and with anti-HA or anti-YFP antibody in (C).