Figures & data
Figure 1. Venn diagrams (drawn approximately to scale) of the data sets used in the census. (A) siRNA libraries and hits, and (B) combined siRNA and pathway analysis results (hits). Large boxed numbers identify the set, and smaller white or red numbers indicate the number of genes or hits in the intersection set. 1: siRNA screen 1; 2: siRNA screen 2; 3: siRNA screen 3; 4: siRNA screen 4; 5: Ingenuity Pathway Analysis (IPA); 6: MetaCore; 7: Pathway Studio (raw hits); and 8: Pathway Studio (manually curated hits). Diagrams with all circles were generated using VennMaster 0.37.5 for calculationsCitation12,Citation13 and then overlaying smooth circles on the VennMaster graphics. When 4 or more sets are being shown as circles, it is not in general mathematically possible to represent them and their intersections graphically to scale with accuracy. VennMaster provides an optimization algorithm that achieves a compromise representation. However, the 8 sets overlap in such a complex way that the fit could be improved by manually changing the circles for sets 6 and 7 to ellipses of approximately the right dimensions. In (A), the larger circle in each case represents the library, and the smaller circle represents validated hits. In (B), some small regions that contain zero hits (colored black) were necessary for graphical purposes.
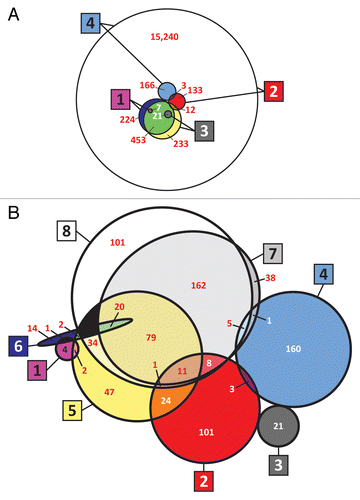
Table 1. Top-ranked autophagy-modulating genes, proteins, and protein complexes
Figure 2. Venn diagrams of the integrated siRNA and pathway analysis results. (A) 739 total genes, proteins, and protein complexes were identified as apparent modulators of autophagy by text mining with our manual curation (yellow circle) and/or siRNA screening (blue circle). The diagram shows the classification of those entities into negative, positive, and dual-potential modulators, and it indicates the tables in Table S1 in which those entities are listed. (B) 385 small molecules were identified by text-mining as modulators of autophagy and categorized in the same manner. Circles were drawn approximately to scale using VennMaster as described in the legend to .
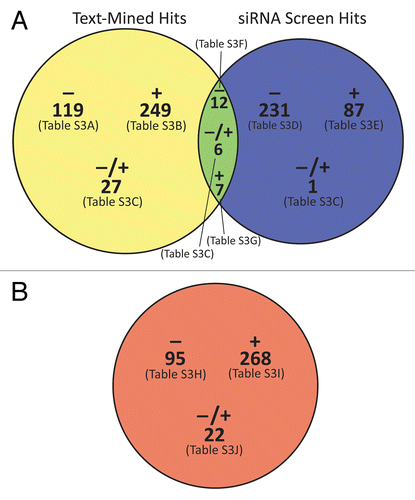
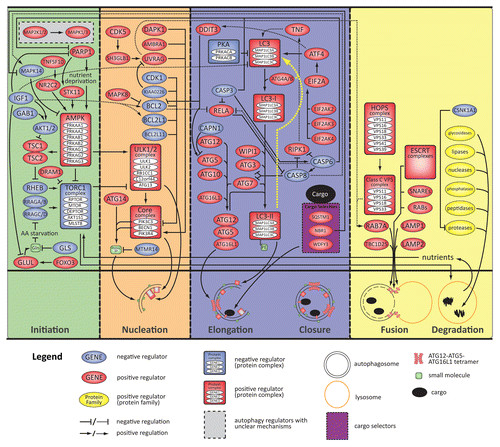
Figure 3. A molecular schematic of the autophagy process based on the information in this census. The top panel shows specific entities (genes, proteins, complexes, and small molecules) associated with the process and, to the extent possible, their specific roles. The middle panel shows a schematic timeline of the 4 stages of macroautophagy from initiation through degradation (colored sections separated by vertical dashed lines). The 3 small molecules depicted are glutamine (Gln), phosphatidylinositol 3-phosphate (PtdIns3P), and phosphatidylethanolamine (PE). Three established cargo selectors include SQSTM1/p62, NBR1, and WDFY3/ALFY. The yellow dotted line in the elongation/closure stage indicates recycling of PE and LC3 by ATG4 following degradation.