Figures & data
Table 1. Phage-encoded effector proteins in E. coli and S. enterica
Figure 1. Schematic of toxins function in eukaryotic cells. Dotted arrows indicate steps in bacterial lysis with phage/toxin release, toxin uptake by host cell and target activity. CTXphi which encodes cholera toxin (CT) does not lyse its bacterial host cell. Black solid arrows or t-bars represent activation or blocking of specified bacterial function by toxin. AB denotes a two subunit protein, subunit A is the catalytic subunit and B is the receptor binding or docking subunit on target cell. AB5 denotes a toxin that contains one A subunit and 5 B subunits. Three examples are given; the A/B diphtheria toxin (DT) and the AB5 toxins CT and shiga toxin (Stx). DT gains direct entry into target cell by pore formation. The second method of toxin uptake is by receptor mediated endocytosis, a mechanism used by CT and Stx. Only the A subunit of DT enters the cell cytosol and along with NAD, ADP-ribosylates Elongation factor 2 blocking protein synthesis. Both CT and Stx are retrograde trafficked to the Golgi apparatus. In CT, the A subunit is dissociated from B subunits in the endoplasmic recticulum (ER) and released into the cytosol where it activates adenylate cyclase increasing cAMP which in turn activates PKA resulting in the CFTR membrane Cl- channel activation. The A subunit of Stx ADP- ribosylates the 28S rRNA ribosomal subunit, thus blocking protein synthesis.
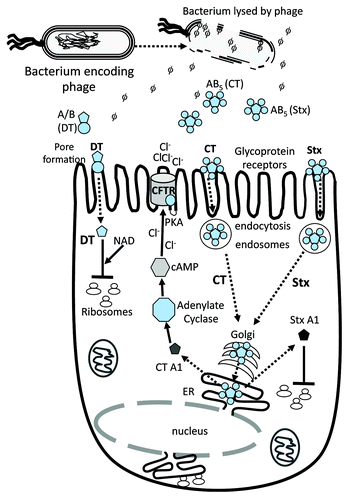
Figure 2. Prophage location on the E. coli O157:H7 strain Sakai genome map. Stars indicate prophages Sp1-Sp18 as named using the convention of.Citation38 Rectangular black box indicates the position of the LEE pathogenicity island. Four letter designations indicate effector genes (red) and toxins (green) identified within prophages and at the LEE island (black).Citation55
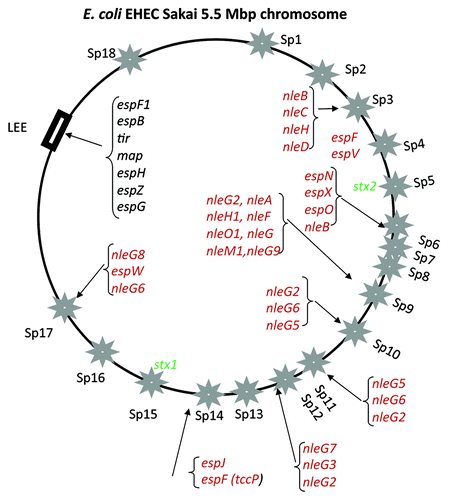
Figure 3. Location of effector genes and stx genes within prophages in E. coli and S. enterica. Partial prophage genomic maps are shown. (A) Terminal region of five lambdoid prophages from E. coli strain Sakai, (B) terminal region of S. enterica Gifsy-1 and Gifsy-2 prophages and (C) central region of the genome of Stx1 (Sp15) and Stx2 (Sp5) prophages encoding stx1AB and stx2AB genes, respectively, shown as blue arrows. Tail fiber genes are represented by white arrows and effector genes are shown as dark gray arrows. Putative additional virulence genes are shown as striped arrows. Black block arrows indicate the prophage site of genomic integration. Arrows in red represent transposase genes.
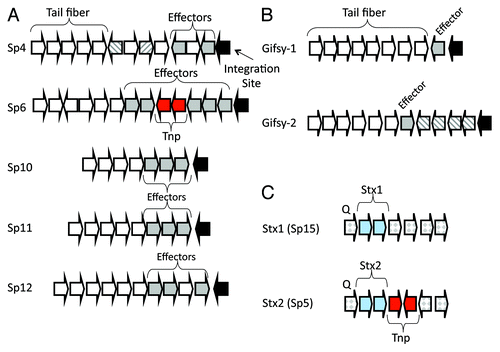
Figure 4. Schematic of the role of different effector proteins in EHEC colonization and pathogenesis. The LEE encoded T3SS is required for intimate binding and adherence of the bacterium to the eukaryotic cell surface which is mediated by the translocation of effector proteins Tir, Map and TccP (EspF). Tir and TccP are required for pedestal formation by directing actin rearrangement within the cell. The NleG family of effector proteins is the largest found in pathogenic E. coli and has been shown to be involved in protein ubiquitination. Many effector proteins are multi-functional such as TccP, which is also involved in cell tight junction disruption and interactions with the nucleus. Prophage-encoded effector genes are shown in red.
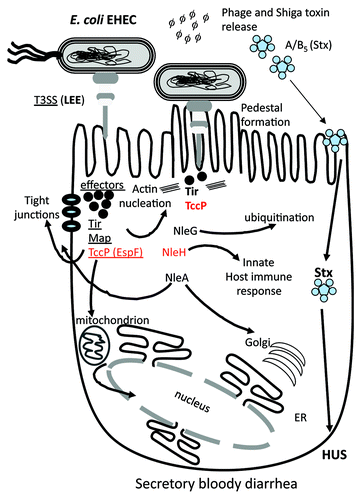
Figure 5. Prophage and pathogenicity island location on the S. enterica serovar Typhimurium genome map. Stars indicate prophages and rectangular black boxes indicate the position of Salmonella pathogenicity islands (SPIs). Four letter designations indicate effector genes identified within prophages (red) and SPIs (black).
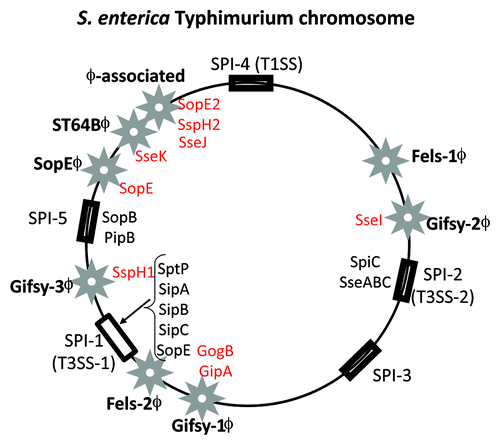
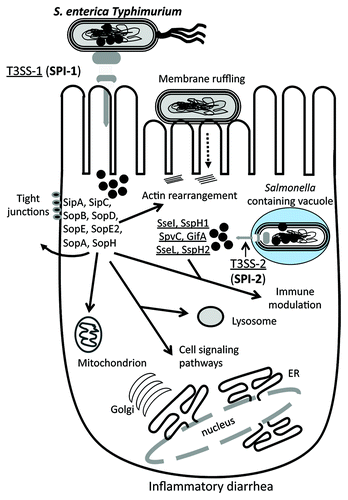
Figure 6. Schematic of the role of different effector protein functions and targets in S. enterica. Salmonella enterica encodes two T3SSs, each within a Salmonella pathogenicity island, SPI-1 and SPI-2. Similar to EHEC effector proteins, a number of effectors are required for intimate binding to the host cell surface, which leads to the uptake into a Salmonella containing vacuole (SCV). Within the SCV, the T3SSs translocates a range of effector proteins into the cytosol that allow for intracellular survival. The S. enterica effector proteins target the tight junctions, mitochondria and lysosomes, as well as several cell signaling pathways. Prophage-encoded effectors are shown in red. See text for details.