Figures & data
Figure 2. C-terminal E-cadherin truncation mutants interact with the cargo-bindng domain of Myosin VI. Coomassie stained gel (A) and the corresponding Myc-immunoblot (B) of a representative in vitro binding experiment. The first two lanes show the E-cadherin tail construct coupled to GSH beads, without (lane 1) and with added myc tagged Myo VI CBD (lane 2). The following 6 lanes were loaded correspondingly with three truncated constructs (NSS, PYD and YYD), as outlined in . Lanes 9–12 are the negative GST controls and lane 13 indicates the molecular mass of the MyoVI CBD construct. The data are representative of three independent experiments.
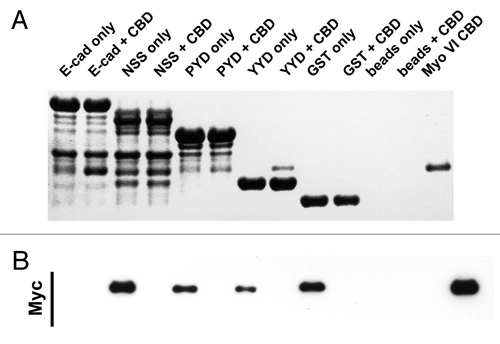
Figure 1. Schema of the E-cadherin cytoplasmic tail and mutants used in this study. The “core” p120 binding siteCitation13 and β-catenin binding site are identified. C-terminal truncations are named for the final (C-terminal) three amino acids in the truncation. The amino acid positions that mark the N- and C-termini of the fragments (numbered from the beginning of the mature protein) are indicated in brackets.
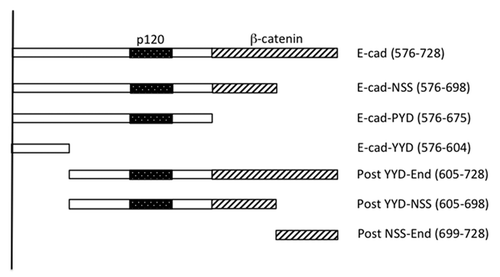
Figure 3. The juxtamembrane cadherin tail is not solely required to bind Myosin VI. Coomassie stained gel (A) and the corresponding Myc-immunoblot (B) of a representative in vitro binding experiment. The first two lanes show the E-cadherin tail construct coupled to GSH beads, without (lane 1) and with added myc tagged Myo VI CBD (lane 2). The following 6 lanes were loaded correspondingly with the three constructs (YYD-end, YYD-NSS and NSS-end) outlined in . Lanes 9–12 are the negative GST controls and lane 13 indicates the molecular mass of the Myo VI CBD construct. The data are representative of three independent experiments.
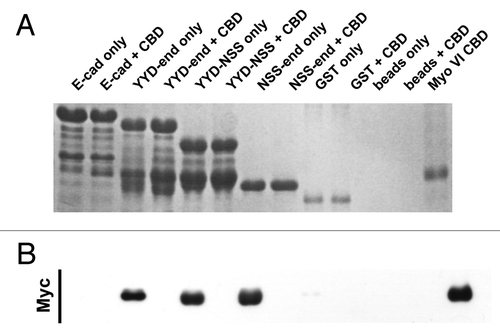
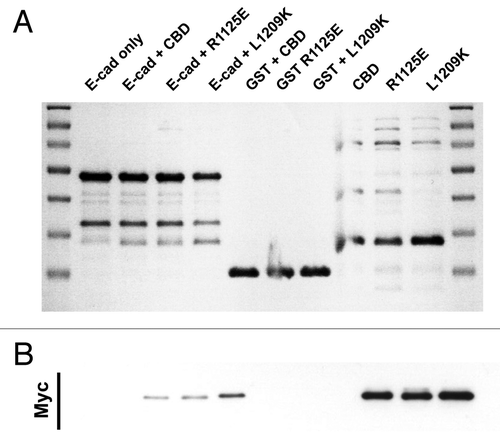
Figure 4. E-cadherin has binding requirements distinct from Dab2. Coomassie stained gel (A) and the corresponding Myc-immunoblot (B) of a representative in vitro binding experiment. The first two lanes show the E-cadherin tail construct coupled to GSH beads, without (lane 1) and with added myc tagged Myo VI CBD (lane 2). In lane 3 and 4 two point mutations of myosin VI (R1125E and L1209K) that inhibit the binding to Dab2 were tested in their ability to bind E-cad. Lanes 5–7 are negative GST controls and lane 8–10 show the three Myo VI CBD constructs loaded alone to indicate their expected molecular mass.