Figures & data
Figure 1. Predicted protein sequence of the sGAI gene of A. awamori (GenBank:JX559863) expressed in S. cerevisiae Y294[ySYAG]. The XYNSEC secretion signal is indicated in bold font. The sequence identified in glucoamylases essential for raw starch hydrolysis was conserved (PL(W-597)YVTVTLPA),Citation19 as well as the second tryptophan (Trp) residue and is double underlined in gray text. The Gp-I region is indicated as text in italics bold. The Cp-I region or Starch Binding Domain is indicated in underlined text.Citation25 The general acid and base catalysts Glu-213 and Glu-434, as well as Tyr-85, Trp-87, Arg-89, Asp-90, Trp-154, Glu-214, Arg-339, Asp-343, Trp-351 sites, which play a role in substrate transition-state stabilization and/or ground-state binding, are indicated in gray text. Likely N-glycosylation sites are underlined by a broken line, although only the first and third sites were found to be glycosylated when expressed in yeast.Citation26
![Figure 1. Predicted protein sequence of the sGAI gene of A. awamori (GenBank:JX559863) expressed in S. cerevisiae Y294[ySYAG]. The XYNSEC secretion signal is indicated in bold font. The sequence identified in glucoamylases essential for raw starch hydrolysis was conserved (PL(W-597)YVTVTLPA),Citation19 as well as the second tryptophan (Trp) residue and is double underlined in gray text. The Gp-I region is indicated as text in italics bold. The Cp-I region or Starch Binding Domain is indicated in underlined text.Citation25 The general acid and base catalysts Glu-213 and Glu-434, as well as Tyr-85, Trp-87, Arg-89, Asp-90, Trp-154, Glu-214, Arg-339, Asp-343, Trp-351 sites, which play a role in substrate transition-state stabilization and/or ground-state binding, are indicated in gray text. Likely N-glycosylation sites are underlined by a broken line, although only the first and third sites were found to be glycosylated when expressed in yeast.Citation26](/cms/asset/6a4ac329-3157-4cd1-a6e7-2078b8de65d9/kbie_a_10922268_f0001.gif)
Figure 2. Glucoamylases production by wild type and recombinant S. cerevisiae strains. (A) Hydrolysis of raw starch appears as clear zones around S. cerevisiae colonies secreting functional glucoamylases; strain (a) Y294 (reference), (b) Y294[yASAG] secreting the native GAI, (c) Y294[ySYAG] secreting the codon-optimized sGAI, were grown for 4 d on agar containing raw starch and then stained with iodine solution. (B) Glucoamylase and α-amylase activities of the strains Y294[yASAG] and Y294[ySYAG]; glucoamylase activity, determined at pH 5.4 and 30°C, was reported as nanokatals per gram dry cell weight (nKat/g DCW), which is the enzyme activity needed to produce 1 nmol of glucose per second per gram dry cell weight. α-amylase activity, detected at pH 5.4 and 50°C, was expressed as Ceralpha Units per gram dry cell weight (CU/g DCW), which is the enzyme activity required to release 1 micromol p-nitrophenyl per min per gram dry cell weight.
![Figure 2. Glucoamylases production by wild type and recombinant S. cerevisiae strains. (A) Hydrolysis of raw starch appears as clear zones around S. cerevisiae colonies secreting functional glucoamylases; strain (a) Y294 (reference), (b) Y294[yASAG] secreting the native GAI, (c) Y294[ySYAG] secreting the codon-optimized sGAI, were grown for 4 d on agar containing raw starch and then stained with iodine solution. (B) Glucoamylase and α-amylase activities of the strains Y294[yASAG] and Y294[ySYAG]; glucoamylase activity, determined at pH 5.4 and 30°C, was reported as nanokatals per gram dry cell weight (nKat/g DCW), which is the enzyme activity needed to produce 1 nmol of glucose per second per gram dry cell weight. α-amylase activity, detected at pH 5.4 and 50°C, was expressed as Ceralpha Units per gram dry cell weight (CU/g DCW), which is the enzyme activity required to release 1 micromol p-nitrophenyl per min per gram dry cell weight.](/cms/asset/71027212-b28d-4601-a3c4-971c60564fb5/kbie_a_10922268_f0002.gif)
Table 1. Glucoamylolytic activity (nKat/DCW) of the engineered S. cerevisiae F2 and F6 strains once grown in YPD broth for 72 h
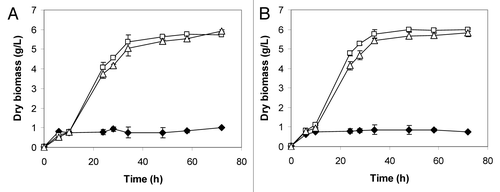
Figure 3. Effect of the temperature on the aerobic growth of S. cerevisiae 27P (♦), F2 (□) and F6 (Δ) incubated at 30°C (A) and 37°C (B) in soluble starch (20 g/L) medium.