Figures & data
Figure 1. Multiple alignment of amino acid sequences (confirmed or predicted mature form) of several plasmodial cysteine hemoglobinases and some closely-related isoforms. Numbers of amino acids are marked at the right side of each of the alignments. Hyphens (-) represent gaps introduced to maximize alignment and asterisks (*) indicate conserved active-site residues. Clear boxes indicate the position of conserved cysteine residues implicated in disulfide bonds (based on the Falcipain-2 structure).Citation7 Shaded boxes indicate the position of a short α-helix (α1) within the N-terminal extension playing a critical role in folding of the mature protein and the C-terminal motif involved in hemoglobin binding. Amino acid sequences of chabaupain-1 (AAP43629); vinckepain-1 (AAL48319); berghepain-1 (XP_677643); yoelipain-1 (XP_729023); vivapain-1 (XP_001615807); falcipain-1 (AAA29578); falcipain-2 (AF282975); falcipain-2´ (AAX77225); falcipain-3 (AAN35746); vivapain-2 (XP_001615274); vivapain-3 (XP_001615273); vivapain-4 (XP_001615272); chabaupain-2 (AAP43630); vinckepain-2 (AAL48319); berghepain-2 (AAL48318); yoelipain-2 (XP_726900) were obtained from GeneBank database (www.ncbi.nlm.nih.gov/genbank/). Papain sequence (AAB02650) was included for comparison.

Table 1. Heterologous expression systems of Papain-like cysteine proteases from human and rodent malaria parasites
Figure 2. Comparison of plasmodial genes for cysteine proteases (●) with highly expressed Escherichia coli genes (٭) corresponding to glycolytic and tricarboxylic acid cycle enzymes. The cluster corresponding to vivipain genes is indicated with dashed lines. The optimized falcipain-2 gene is also indicated with a star. CAI values and global A+T content were calculated using the CAIcal server (http://genomes.urv.es/CAIcal/).Citation27
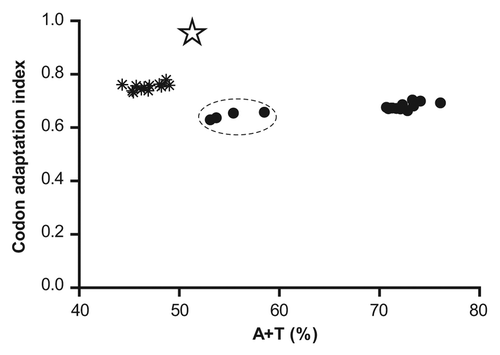
Figure 3. Comparison of several eukaryotic systems (hosts) for the expression of plasmodial genes for cysteine proteases (P. pastoris: Pichia pastoris; S. cerevisiae: Saccharomyces cerevisiae; T. ni: Trichoplusia ni; S. frugiperda: Spodoptera frugiperda; C. griseus: Cricetulus griseus; H. sapiens: Homo sapiens). The mean for each group is indicated by a horizontal line. The cluster corresponding to vivipain genes is indicated with dashed lines.
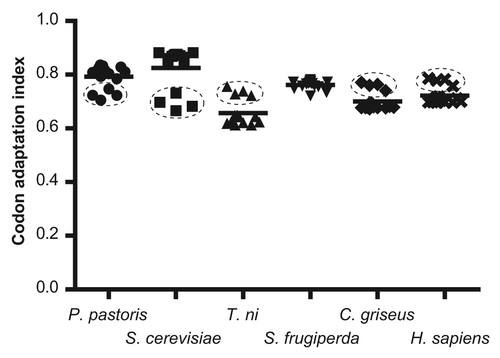
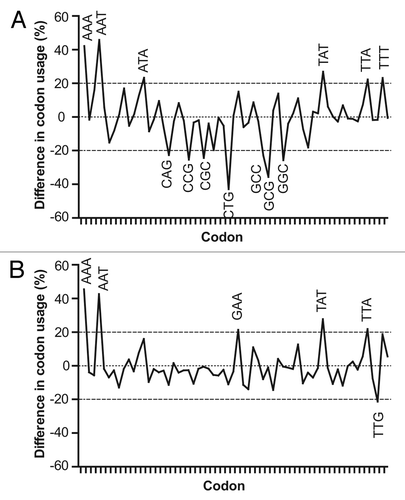
Figure 4. Comparative profile of relative codon frequencies in plasmodial cysteine protease, E. coli (A) and P. pastoris (B) genes. Differences in frequency (per thousand) of codon usage (FCUPLASMODIUM – FCUHOST) were calculated for each codon, using codon composition from plasmodial cysteine protease genes and information contained in codon usage tables for E. coli and P. pastoris, obtained from Codon Usage Database.Citation30 Those codons showing modular differences ≥ 20% are indicated.