Figures & data
Figure 1. Structure of PP2A (PDB ID: 3dw8,Citation38). The quaternary protein structure is shown, composed of the catalytic subunit Cα, the 65 kDa regulatory subunit Aα, and the 55 kDa regulatory subunit Bα. The two manganese atoms present in the catalytic subunit are also shown.
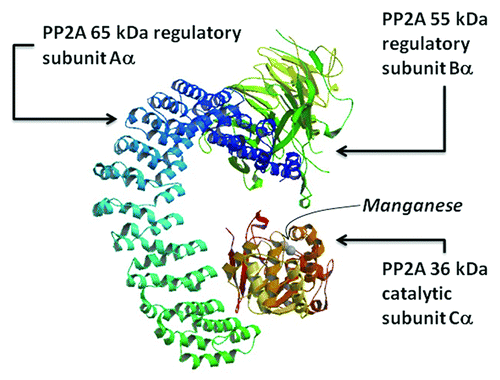
Table 1. Comparison of purification results for PP2A obtained from different sources, showing the enzyme quantity, activity and inhibition by OA
Figure 2. Activity of PP2ACα purified from insect larvae. Activity was determined after 1, 2 or 12 mo after purification by the 4-MUP method. The enzyme was stored in 50 mM TRIS-HCl, pH 7.2, or in 50 mM TRIS-HCl pH 7.2 and 50% glycerol. The enzyme retained 100% of its activity after 2 mo, when stored in 50 mM TRIS-HCl, pH 7.2, and lost approximately 30% of its activity after a year in the same conditions. On the other hand, there was a decrease in enzymatic activity after 1 mo when the PP2ACα was dissolved in 50 mM TRIS-HCl pH 7.2 and 50% glycerol. These decrease in enzyme activity was more pronounced after 2 mo and almost no activity was observed after one year. AUF, arbitrary fluorescence units; MIN, minutes.
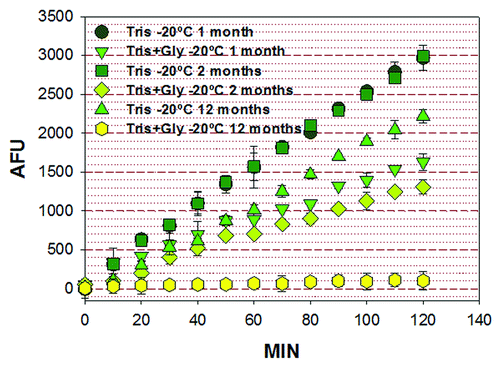
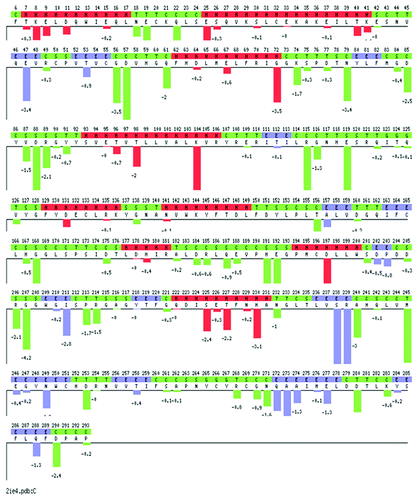
Figure 3. Plot of the sequence optimality score (sum of negative ΔΔGs) for each position in the PP2ACα sequence obtained with the PoPMuSiC (Prediction of Protein Mutant Stability Changes) server. The ΔΔG predicts the change in folding free energy upon mutation. ΔΔG values are given in kcal/mol. Red: Helices, blue: β-strands, and green: turns and coils. The predicted stability changes for each possible mutation, and the sequence optimality score computing the sum of negative ΔΔGs for each position in the sequence can be accessed at the PoPMuSiC server introducing the PDB ID: 2ie4.