Figures & data
Figure 1. Schematic representation illustrating the activation of SUMO by an E1 enzyme. (A) Conjugation of SUMO to a substrate requires the initial activation of SUMO by the SUMO-activating enzyme (E1), a heterodimer of Aos1 and Uba2. The E1 enzyme uses ATP to activate the carboxyl terminus of SUMO as a high-energy anhydride (SUMO-AMP). This step is termed adenylation. The E1 catalytic cysteine residue then attacks the adenylated SUMO to form a thioester intermediate, referred to as thioesterification. The activated SUMO molecule is then transferred through a trans-thioesterification to a cysteine residue in the SUMO-conjugating enzyme (E2; Ubc9). Finally, the E2-bound SUMO molecule attacks a substrate either in the presence or in the absence of an SUMO-E3 ligase; this process is termed amidolysis. (B) The domain organization of mouse ubiquitin E1 (Uba1; the top bar) and SUMO-E1 (a heterodimer of Aos1 and Uba2 subunits; the middle bars). A predicted structure of the Aos1-Uba2 chimeric recombinant protein is also illustrated and designated as mAU (the bottom bar). The numbers represent residues of each polypeptide. The catalytic cysteine residues at 632, 173, and 425 amino acid residues in Uba1, Una2, and mAU, respectively, are also indicated as vertical lines.
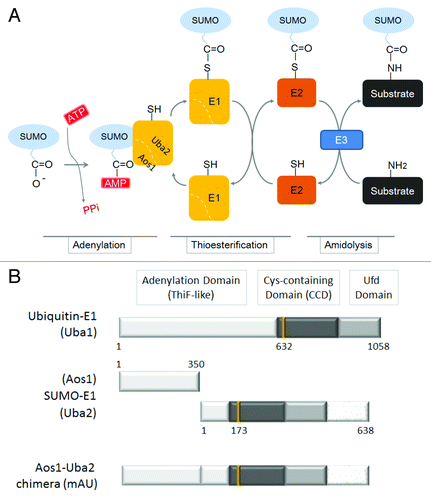
Figure 2. Construction of the mAU baculovirus/insect expression vector and the in situ SUMOylation assay using the recombinant His6-mAU. (A) Mouse AU was amplified and cloned into NotI and XhoI sites of the multi-cloning site of pFastBac-HT-C. The pFastBac-HT-C vector plasmid is designed to express inserted gene fragments in the multi-cloning site as fusion proteins with an N-terminal 6 × His-tag sequence (His6) followed by the Tobacco Etch Virus (TEV) protease cleavage site. Thus, 45 residues including the His6 and TEV recognition/cleavage regions were present in our construct. These additional residues were considered to not influence SUMO-E1 enzymatic activity of mAU. The EcoRI site inserted between the Aos1 and Uba2 fragments is indicated. ATG, start site; Six × His, hexa-histidine tag sequence; TEV, TEV-protease recognition site and cleavage site; MCS, multi-cloning sites. (B) Schematic representation of His6-mAU recombinant protein. The numbers show amino acid residues. There are two amino acid residues between Aos1 and Uba2, glutamic acid (E), and phenylalanine (F), which were derived from the linker fragment of the EcoRI site (see the plasmid map in [A]). (C) Comparison of bacteria expressed and baculovirus-insect cell-expressed mAU proteins. Total lysate from either GST-mAU-expressing E. coli or His6-mAU expressing baculovirus-insect cells was subjected to immunoblot analysis using an anti-GST or anti-His6 antibody, respectively. Positions of full-length GST-mAU and His6-mAU recombinant proteins are indicated by arrows. Protein size markers are shown on the left. (D) Application of His6-mAU recombinant protein to the in situ SUMOylation assay.Citation29,Citation30 HeLa cells were permeabilized with 0.01% digitonin followed by 4% paraformaldehyde fixation. Twenty μl of the in situ SUMOylation buffer containing Ubc9, GFP-SUMO-1, and Mg2+-ATP, supplemented with (+, left column) or without (−, right column) the His6-mAU recombinant protein. After incubation for 20 min at 18 °C, GFP (upper panel) and DAPI (bottom panel) signals were observed by fluorescence microscopy. Bars indicate 10 μm.
![Figure 2. Construction of the mAU baculovirus/insect expression vector and the in situ SUMOylation assay using the recombinant His6-mAU. (A) Mouse AU was amplified and cloned into NotI and XhoI sites of the multi-cloning site of pFastBac-HT-C. The pFastBac-HT-C vector plasmid is designed to express inserted gene fragments in the multi-cloning site as fusion proteins with an N-terminal 6 × His-tag sequence (His6) followed by the Tobacco Etch Virus (TEV) protease cleavage site. Thus, 45 residues including the His6 and TEV recognition/cleavage regions were present in our construct. These additional residues were considered to not influence SUMO-E1 enzymatic activity of mAU. The EcoRI site inserted between the Aos1 and Uba2 fragments is indicated. ATG, start site; Six × His, hexa-histidine tag sequence; TEV, TEV-protease recognition site and cleavage site; MCS, multi-cloning sites. (B) Schematic representation of His6-mAU recombinant protein. The numbers show amino acid residues. There are two amino acid residues between Aos1 and Uba2, glutamic acid (E), and phenylalanine (F), which were derived from the linker fragment of the EcoRI site (see the plasmid map in [A]). (C) Comparison of bacteria expressed and baculovirus-insect cell-expressed mAU proteins. Total lysate from either GST-mAU-expressing E. coli or His6-mAU expressing baculovirus-insect cells was subjected to immunoblot analysis using an anti-GST or anti-His6 antibody, respectively. Positions of full-length GST-mAU and His6-mAU recombinant proteins are indicated by arrows. Protein size markers are shown on the left. (D) Application of His6-mAU recombinant protein to the in situ SUMOylation assay.Citation29,Citation30 HeLa cells were permeabilized with 0.01% digitonin followed by 4% paraformaldehyde fixation. Twenty μl of the in situ SUMOylation buffer containing Ubc9, GFP-SUMO-1, and Mg2+-ATP, supplemented with (+, left column) or without (−, right column) the His6-mAU recombinant protein. After incubation for 20 min at 18 °C, GFP (upper panel) and DAPI (bottom panel) signals were observed by fluorescence microscopy. Bars indicate 10 μm.](/cms/asset/4d952b0e-043d-4f91-a083-581108a6f38c/kbie_a_10927544_f0002.gif)
