Figures & data
Figure 1. Eukaryotic translation of mRNA. Methylated 5′ cap, 5′m; translation initiation site, TIS (a motif that also includes the start codon); stop codon; ribosomal subunits, ovals; protein-coding ORF, green rectangle; protein, squiggle; polyA, AAA. The asterisk denotes factors that complete the ribosomal preinitiation complex.
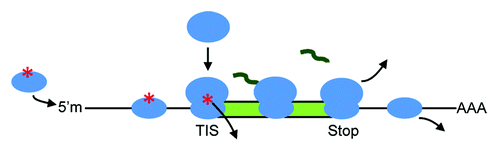
Figure 2. The TIS sequence and eIFs mediate initiation of translation. (A) 43S preinitiation complex (40S, eIFs 1, 1A, 2, 3, 5, and initiator Met-tRNA) recognizes a TIS sequence (in this example, GCCACCAUGGC with start codon underlined). (B) eIF5B is loaded while others detach. This allows the joining of the 60S ribosomal subunit to form the 80S complex. After shedding the remaining eIFs, (C) protein elongation begins. Initiator Met-tRNA, red L; eIFs, white circles; bases represent the TIS sequence.
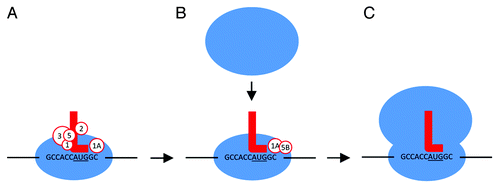
Figure 3. Schematic of leaky scanning and reinitiation. 43S complex loaded with tRNA plus initiation factors and primed for initiation, small blue oval with red asterisk; 40S subunit not competent for initiation, small blue oval without asterisk; 60S subunit, large blue oval; ORFs, rectangles; squiggle represents ongoing translation.
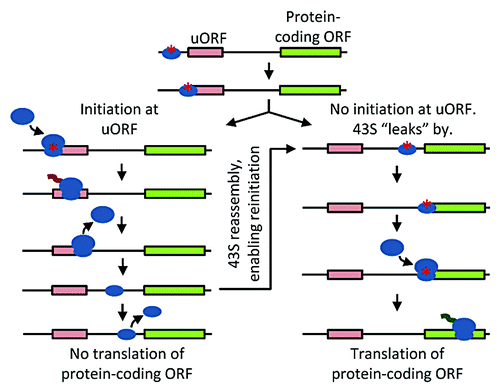
Figure 4. Schematic of protein translation tuning achieved by specifying TIS sequences of upstream and protein-coding ORFs. Because of the possibility of leaky scanning, ribosomes can take three possible paths (thick-lined arrows). By varying the three TIS bases (NNN) preceding the start codon of uORFs or protein-coding ORFs, the distribution of ribosomes over these paths is controlled. In doing so, one controls the flux of ribosomes that both reach and translate the protein of interest. Additional control can also be achieved by using multiple uORFs in series and non-AUG start codons (not shown).
Figure 5. Control of protein translation levels. (A) Varying GFP expression levels achieved in different cell lines using MoMLV retroviral vectors equipped with engineered RNA leaders. Leaders utilizing different TIS sequences and uORFs were used to generate lower expression levels. Leaders with varying TIS sequences and no uORFs were used to generate higher expression levels. (B) Lentiviral expression of GFP in HCT-116 cells normalized to an internal RFP reference. (C) Immunoblot of retroviral expression of DHFR-BFP-RasG12V in NIH-3T3 cells with (+) and without (-) trimethoprim, the small molecule inducer of DHFR fusion protein stability. Sequences of the RNA leaders are given with uORFs in red bold type (including stop codons); start codons are underlined; the first base of the start codon encoding a protein of interest is indicated by the +1 base position. Error bars represent standard deviations from triplicate experiments.
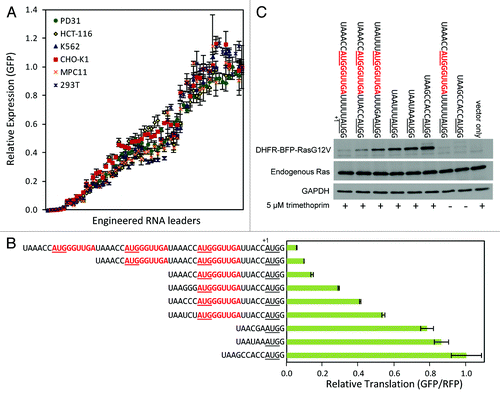
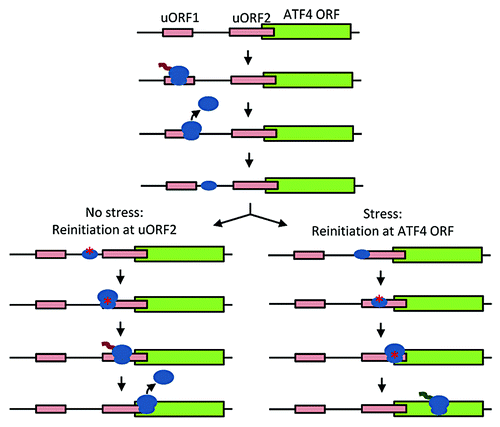
Figure 6. Schematic of stress-induced translation of ATF4. After translating uORF1, under non-stressed conditions, the 40S subunit (small oval) quickly reloads with eIF2-GTP, Met-tRNA, and other eIFs (red asterisk) and reinitiates translation at uORF2. Under stress, the 40S subunit is not reloaded until later, reinitiating translation at ATF4.