Figures & data
Figure 1. 3D invasions of NMuMG and 4T1 cells are increased by conditioned media from both mammary epithelial cells and D1 mesenchymal stem cells. Migration of NMuMG (A) and 4T1 cells (B) following a 5-d incubation with CMs derived from 4T1, NMuMG, and the mesenchymal stem D1 cells were quantified using 3D migration assays (see Materials and Methods section for details). Serum-free and 10% FBS–DMEM media were used as negative and positive controls, respectively. Representative microphotographs of cells migrating out of the wells for each condition were captured following the 5-d incubation. In experiments repeated at least 3 times, the cells moving toward different control and CMs were counted. Data were analyzed by one-way ANOVA and differences between treatment groups tested using the Student Newman–Keuls post-hoc test. *P < 0.05 compared with negative control (0% FBS).
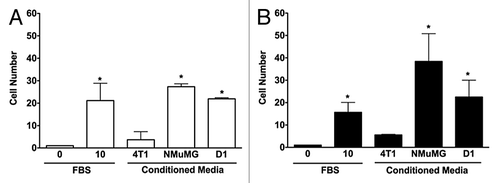
Figure 2. D1 mesenchymal stem cell conditioned media promotes the invasion of 4T1 cells but not NMuMG cells. Invasion of NMuMG and 4T1 breast cells placed in the upper chamber of Matrigel®-coated transwells toward a bottom chamber filled with conditioned media (1:1 dilution) collected from 4T1, NMuMG, or mesenchymal stem D1 cells was evaluated following a 24-h incubation. Representative microphotographs of NMuMG and 4T1 cells labeled with the vital nuclear dye Hoechst that migrated through Matrigel®-coated transwell membranes toward lower compartments filled with each treatment are presented (A). All microphotographs were taken in the same conditions of fluorescence and magnification (bar scale = 100 μm, lower right), converted to gray scale and inverted. Serum-free (0% FBS) and 10% FBS media were used as negative and positive control, respectively. The invasion was evaluated by counting the number of NMuMG (B) and 4T1 (C) cells on at least 5 representative pictures for each condition. The number of cells was then normalized to the surface area of the transwell membrane. Data were analyzed by one-way ANOVA and differences between treatment groups tested using the Student Newman–Keuls post-hoc test. *P < 0.05 compared with cells migrating toward media (0% FBS) alone.
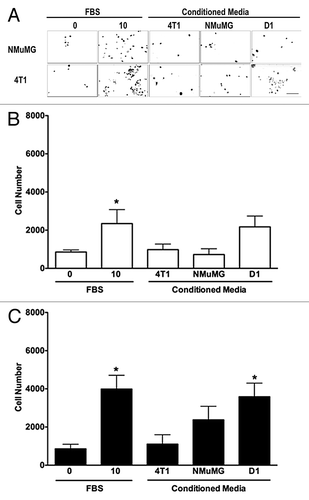
Figure 3. D1 mesenchymal stem cell conditioned media contains higher CCL-5 and CCL-9 concentrations than conditioned media collected from NMuMG mammary epithelial and 4T1 tumor cells. Higher concentrations of CCL5 and CCL9 were detected in CM collected from mesenchymal stem cells (D1) (A), than in CM obtained from mammary epithelial cells (NMuMG) (B) and mammary tumor cells (4T1) (C) using cytokine protein arrays. This increase was semi-quantified (D) in CMs collected from D1 (gray bars), and in 4T1 (black bars) cells. Both CCL5 AND CCL9 expressions were very low in NMuMG conditioned media (below the detection limit, not shown). Data were analyzed by one-way ANOVA and differences between treatment groups tested using the Student Newman–Keuls post-hoc test. *P < 0.05, **P < 0.01 compared with 4T1 conditioned media.
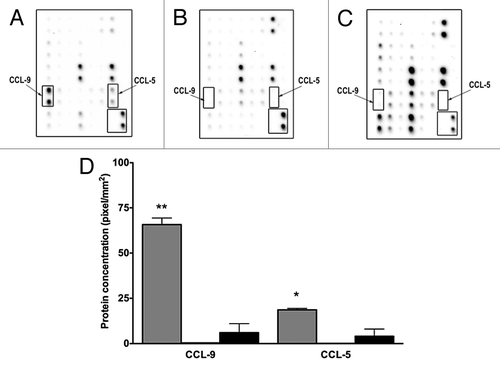
Figure 4. D1 mesenchymal stem cell conditioned media stimulated the mRNA expressions of CCR1 and CCR5 in 4T1 cells but not in NMuMG cells. The mRNA expression for CCR1, CCR3 and CCR5 was evaluated and quantified by RT-PCR in RNA collected from NMuMG (A, C, and E) and 4T1 (B, D, and F) cells following a 3 h incubation with controls and CMs from 4T1, NMuMG and D1 mesenchymal stem cells, respectively. The primer sequences are listed in . Serum-free (0% FBS) and 10% FBS media served as negative and positive control, respectively. Using experiments repeated at least three times, CCR1 (C and D) and CCR5 (E and F) mRNA expressions in NMuMG (C and E) and 4T1 (E and F) cells normalized to loading control (GAPDH) mRNA expression were quantified. The expression of CCR3 mRNA was unaffected regardless of the treatments (P > 0.05, not shown). Results expressed as fold increase compared with 0% FBS control were analyzed by one-way ANOVA and differences between treatment groups tested using the Student Newman–Keuls post-hoc test. *P < 0.05 compared with negative control (0% FBS).
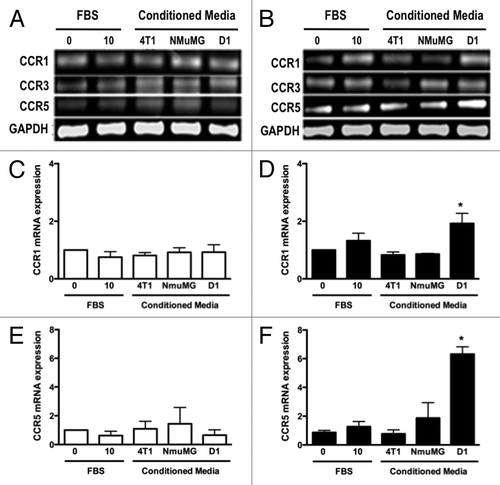
Figure 5. D1 mesenchymal stem cell conditioned media increased cell surface expression of CCR1 and CCR5 receptors in 4T1 cells. Representative flow-cytometry histograms that depict the CCR1 (A) and CCR5 (B) receptors expressed on the cell surface of 4T1 cell following a 24 h incubation in the presence of control media (dotted gray line) or D1CM (solid dark line). Staining with the secondary antibody alone (Texas red conjugated anti-goat antibody) is depicted as solid black area. These results are representative of at least three separate experiments. Results indicate that incubation with D1CM led to a limited and a larger cell surface expression of CCR1 and CCR5, respectively.
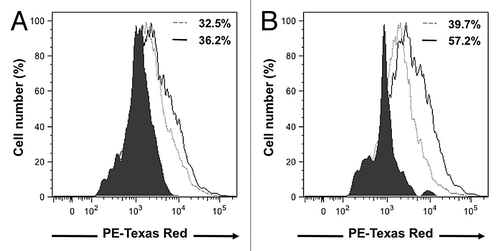
Figure 6. The inhibitor of CCR1 and CCR5 Met-CCL5 inhibits 4T1 cells invasion toward D1 mesenchymal stem cell conditioned media. The number of 4T1 cells migrating through uncoated and Matrigel®-coated transwell chamber was used to evaluate the effects of increasing concentrations of met-CCL5, an inhibitor of both CCR1 and CCL5 on the invasion promoted by mesenchymal stem D1 cell CM (for details see materials and Methods section). Serum-free (0% FBS) and 10% FBS media serve as negative and positive control, respectively. The numbers of migrating 4T1 cells normalized to the surface area of the transwell membrane are presented. Data were analyzed by one-way ANOVA and differences between treatment groups tested using the Student Newman–Keuls post-hoc test. *P < 0.05, **P < 0.01, ***P < 0.001 compared with 0% FBS control; #P < 0.05, ##P < 0.01, ###P < 0.001, compared with migration of 4T1 cells toward D1CM alone.
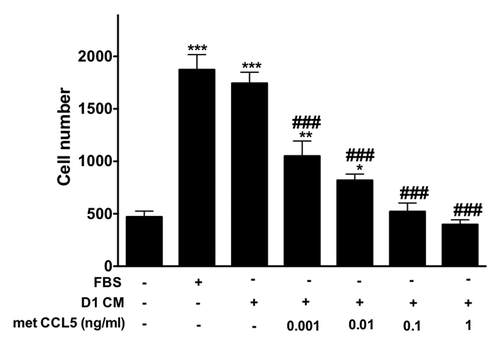
Figure 7. The increased 4T1 cell invasion promoted by CCL5 and CCL9 alone was prevented through pre-treatment with the MMP inhibitor GM6001. Briefly, 4T1 tumors cells were pre-incubated without (black bars) or with GM6001 (1 μM, hachured bars) for 30 min prior to invasion assays. Invasion assays were conducted using transwell chambers (for details see Materials and Methods section). The bottom chamber was filled with either media alone, CCL5 (12.5μM), CCL9 (12.5μM) the combination of both or DICM. The numbers of migrating 4T1 cells normalized to the surface area of the transwell membrane are presented. Data were analyzed by one-way ANOVA and differences between treatment groups tested using the Student Newman–Keuls post-hoc test. *P < 0.05, **P < 0.01, ***P < 0.001 compared with migration of 4T1 cells toward 0% FBS alone.
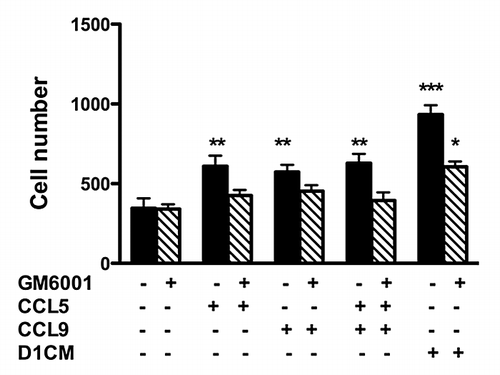
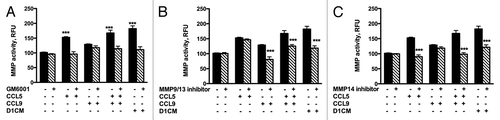
Figure 8. D1 mesenchymal stem cell CM, CCL5, and CCL9 promoted the activities of MMPs including MMP9/13 and MMP14. MMP activities of supernatants collected from 4T1 cells incubated 24 h with either media alone, CCL5, CCL9, or D1CM were freeze-dried and concentrated to 1/10 of their initial volume were assayed according to the manufacturer’s recommendations (see Materials and Methods for details). To determine whether the activities of MMP9/13 or MMP14 were altered by either CCL5, CCL9, the combination CCL5–CCL9 or D1CM, specific inhibitors of MMP9/13 (10 nM) and MMP14 (50 nM), along with the general MMP inhibitor GM6001 (10 nM) were added to the reaction. Data are expressed in relative fluorescence unit (RFU). Data were analyzed by one-way ANOVA and differences between treatment groups tested using the Student Newman–Keuls post-hoc test. ***P < 0.001 compared with MMP activities of 4T1 cells incubated with 0% FBS alone.