Figures & data
Figure 1. CXCR7 expression results in removal of CD31/PECAM-1 from cell-cell junctions. pLEC were infected with Trans or Trans+CXCR7. (A) At 20 h post-infection cultures were fixed and stained by IFA for PECAM-1 (green), CXCR7 (red), and DAPI. The white star marks the nucleus of an internal control CXCR7 negative cell in contrast to CXCR7+ cell denoted by white triangle. Scale bar is 30 µm. (B) Identical cultures were lysed and analyzed by western blot for GAPDH and CD31/PECAM. Data are representative of three replicate experiments. Membranes were then reprobed for HA to verify adenovirus transduction efficiency. The star (*) denotes a non-specific band commonly detected by HA antibody in pLEC lysates. As with many proteins containing multiple transmembrane domains, HA-CXCR7 in boiled lysates appears as a smear due to aggregation of the hydrophobic domains.
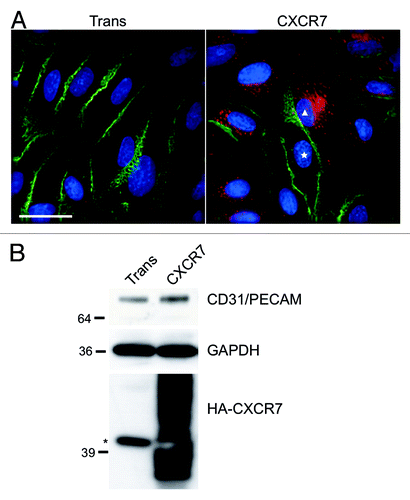
Figure 2. CXCR7+ EC display altered morphology and lack contact inhibition. pLEC were infected with Trans+CXCR7. At 20 h post-infection cultures were fixed and stained by IFA for PECAM-1 (green), CXCR7 (red), and DAPI. (A) A field of interest was selected and a z-stack series of 40 images with 0.1 µM step size was photographed at 100X magnification. The image was subjected to deconvolution analysis. Nuclei of CXCR7 negative cells are marked with white stars, the nucleus of the CXCR7+ cell is marked with a white triangle. Scale bar is 20 µm. A region of interest (white box) containing both CXCR7 staining and junctional CD31 staining was extracted and (B) subjected to 3-dimensional analysis. White dashed lines denote the region in the x-y plane containing junctional CD31 staining.
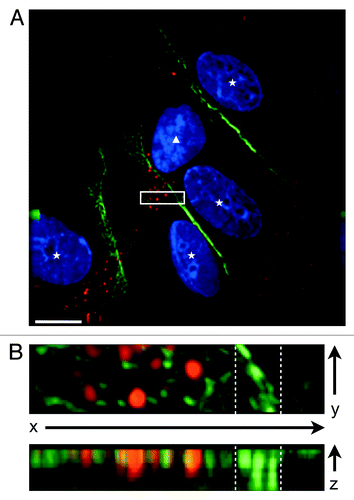
Figure 3. CXCR7+ EC are defective in barrier formation. pLEC were infected with Trans or Trans+CXCR7. (A) At 20 h post-infection cells were trypsinized and replated onto ECIS arrays with or without SDF-1/CXCL12 at 10 ng/ml and allowed to attach for 25 h. Impedance readings at 4000 Hz were taken every 8 s throughout the timecourse. Individual wells were normalized to impedance at t = 0 and duplicate wells were averaged to create the impedance curves. Data are representative of duplicate wells from three independent experiments. (B) At the time of ECIS seeding, a subset of cells was reserved for analysis of CXCR7 and CXCR4 expression by flow cytometry. Necrotic cells were excluded from the analysis via propidium iodide staining and scatter characteristics. (C) Duplicate multi-well plates seeded identically to ECIS arrays were fixed at 25 h post-seeding and stained for CXCR7 (red), PECAM-1 (green), and DAPI and analyzed by deconvolution microscopy.
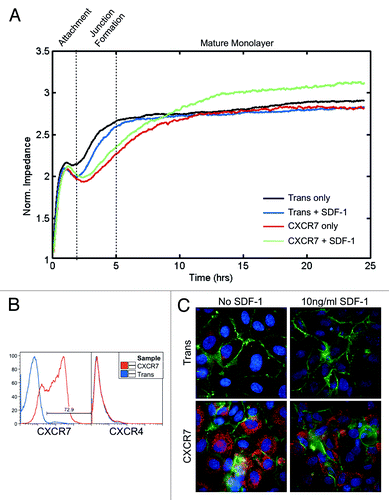
Figure 4. CXCR7+ EC display increased micromotion by ECIS. (A) Extracted impedance readings from ECIS traces scaled to 2 h by 100 ohms. Curves shown are impedance traces of pLEC infected with Trans or Trans+CXCR7 and treated with media only (Unstimulated, top 3 panels) or 10 ng/ml SDF-1/CXCL12 (bottom three panels). Symbols indicated for each experiment correspond to the data points shown in (C). (B) Overlayed time differential (left) and binned histograms of time differential data (right) for Trans (black, overlayed) and Trans+CXCR7 (blue, underlayed) corresponding to the same panels in (A). (C) Quantitation of the standard deviation of impedance fluctuations in Trans or Trans+CXCR7 cultures with or without 10 ng/ml SDF-1/CXCL12. Analysis includes duplicate wells from three independent experiments (black data points) and the average for each condition (gray bar).
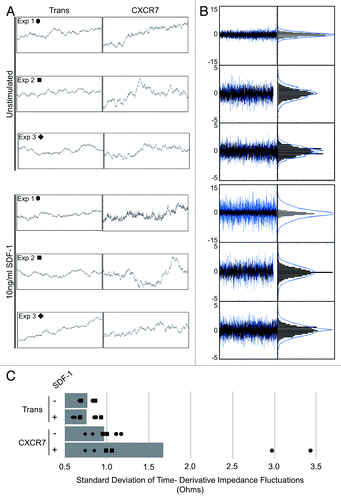
Figure 5. CXCR7 expression does not affect angiogenesis. pLEC were infected with Trans or Trans+CXCR7. At 20 h post-infection cells were trypsinized and replated onto matrigel in complete medium or serum-free medium with or without SDF-1 at 50 ng/ml. Tubule formation was analyzed at 24 h post-plating by (A) light microscopy and (B) quantitation of branch points in 3–5 wells per condition from four independent experiments. P > 0.8 for Trans vs. CXCR7 in all conditions. P > 0.2 for SFM vs. SDF-1 for both Trans and CXCR7.
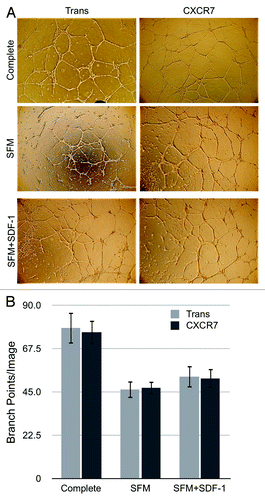
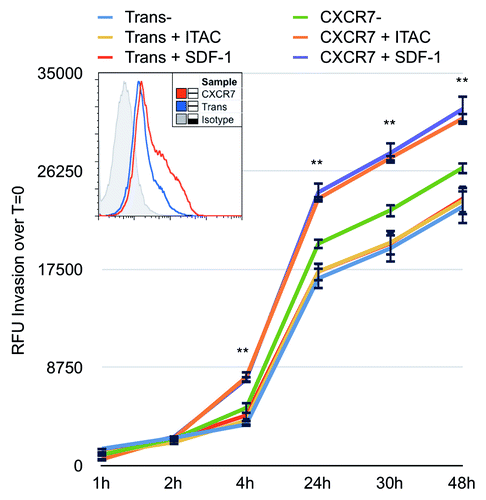
Figure 6. CXCR7 expression enhances EC invasion toward CXCR7 ligands. iLEC were infected with Trans or Trans+CXCR7. At 20 h post-infection cells were labeled with Calcein-AM dye, trypsinized, and transferred to matrigel-coated Fluoroblok™ invasion plates with ITAC/CXCL11 at 50 ng/ml, SDF-1/CXCL12 at 50 ng/ml, or no ligand in the bottom chamber. Invasion was measured via fluorescence accumulation in the bottom chamber at the indicated timepoints. Individual wells were normalized to fluorescence intensity at t = 0 and RFU increases were averaged for each condition. n = 16 wells per condition and data are representative of three independent experiments. **P < 0.001 for both CXCR7+SDF-1 and CXCR7+ITAC compared with unstimulated Trans control. A subset of cells was stained prior to seeding for HA-CXCR7 by flow cytometry to control for adenovirus transduction efficiency (histogram, inset).