Figures & data
Figure 1 Proteolytic processing of APP. A schematic depiction of APP proteolytic processing. APP is a type I transmembrane protein. The Aβ peptide domain is located partially within the membrane spanning hydrophobic segment. In amyloidogenesis, the β-secretase recognition site is cleaved by BACE1 to a soluble fragment N-terminal fragment sAPPα, a C-terminal fragment CTFβ. The latter is then cleaved intramembranously by γ-secretase to yield Aβ and the APP intracellular domain (AICD). In the non-amyloidogenic pathway, α-secretases cleave at a site within the Aβ peptide sequence. This cleavage essentially disrupts the β-secretase recognition site. α-secretases cleavage yields the soluble N-terminal sAPPα, and the C-terminal CTFα. The latter can also be cleave by γ-secretase to yield a non-toxic 3 kDa fragment known as p3, and the APP intracellular domain (AICD).
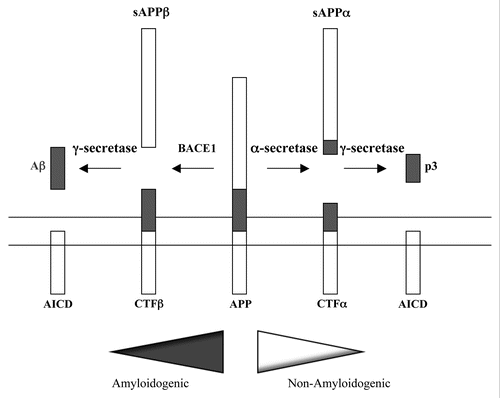
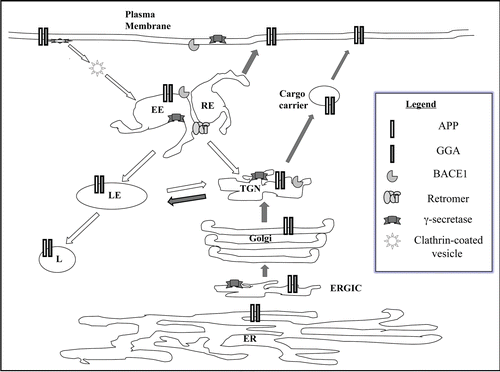
Figure 2 Trafficking itinerary of APP and BACE1. A schematic diagram of the trafficking itinerary of APP and BACE1. Both proteins have very similar itinerary and can be found in compartments of the exocytic pathway and endocytic pathway. Locations of general components of membrane traffic known to affect APP/BACE1 traffic, such as gamma-ear-containing ARF-binding (GGA) proteins and the retromer complex are also shown. Although presenilins are largely localized to the ER-Golgi region at steady state, these and other components of the γ-secretase complex also have widespread presence. Note that this is a general scheme which does not include detail depiction of neuronal domains such as axons and dendrites. Dark arrows indicate exocytic or anterograde transport and white arrow endocytic or retrograde trafficking. ER, endoplasmic reticulum; ERGIC, ER-Golgi intermediate compartment; TGN, trans-Golgi network; EE, early endosome; RE, recycling endosome; LE, late endosome; L, lysosome.