Figures & data
Figure 1 Splicing variant-dependent susceptibility to the degradation process of SDF-1 in blood. Large squares correspond to the ordered structure of the protein. Disordered ends are represented by thinner shapes. The regions of the protein resistant to proteolysis are marked in blue, while parts colored in red represent endings susceptible to the degradation process. The lowest row of words describes the contribution of particular exons to the final structure of the SDF-1 protein. The scissors symbolize the sites of proteinase attack. The large gray arrow in the center of the figure points to the phenomenon of C-terminus degradation prevention by an attachment of a fourth exon resistant to proteolysis during the splicing process.
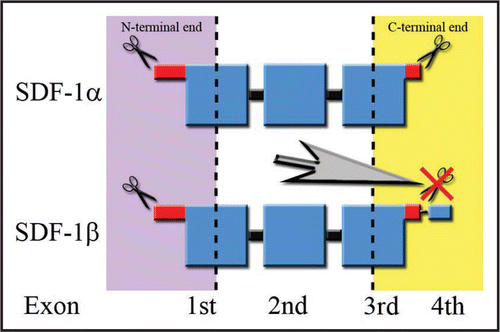
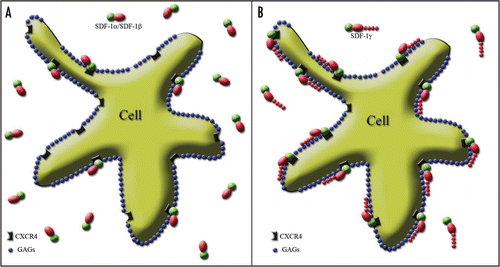
Figure 2 Splicing variant-dependent strength of SDF-1 binding to CXCR4 on the cell surface. The cell membrane is covered by negatively charged GAGs shown as blue spots. The active part of the SDF-1 molecule binding to the receptor is marked in green. Red shapes represent the positively charged part of the SDF-1 molecule stabilizing the process of receptor activation through their interaction with the negatively charged GAGs. (A) The red oval reflects the “standard” positive charge characteristic of all SDF-1 molecules. It is similar for SDF-1α and SDF-1β and is strong enough only for short term receptor binding. Thus, only a limited number of SDF-1 molecules interact with the receptor at the same time, while most of them are in an unbound state. (B) In contrast, the addition of a long, strongly positively charged product of the fourth exon in the case of the SDF-1γ isoform, which is represented in the figure by a “tail” of red spots, facilitates a very strong and long-term stabilization of SDF-1 binding to CXCR4. It results in the persistency of SDF-1 activity which is shown by widespread binding of the SDF-1 molecule to its cognate receptor.