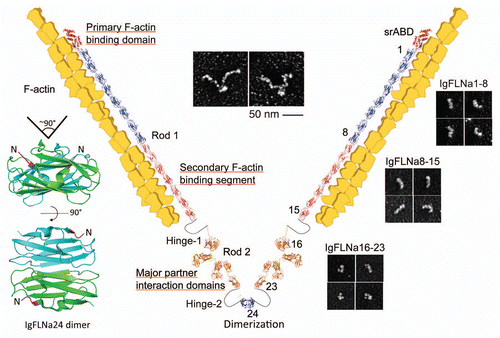
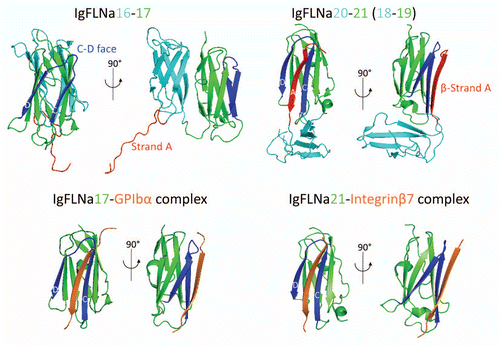
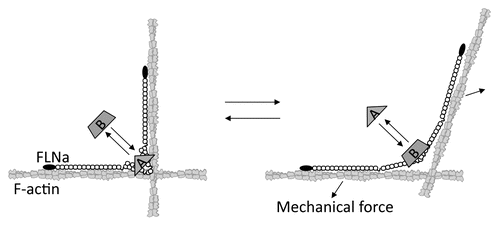
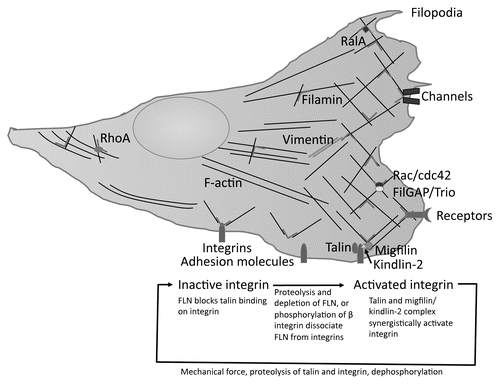
Figures & data
Table 1 Filamin binding partners involved in cell adhesion, spreading and migration