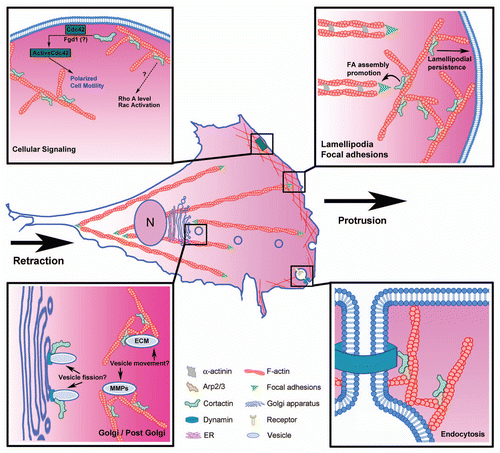
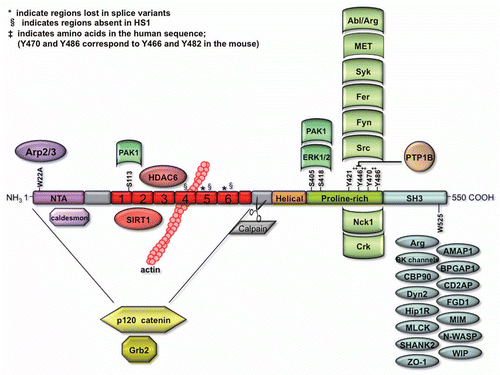
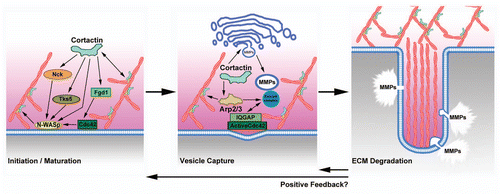
Figures & data
Table 1 Table of cortactin binding partners