Figures & data
Figure 1 eIF4E expression and phosphorylation in breast cancer tissue. Equal protein concentrations from 10 breast cancer tissue samples were separated by SDS-PAGE and western blots probed with antibodies to eIF4E and serine 209 phosphorylated eIF4E. Figure representative of two independent blots. Quantitation shows the relative ratios of phosphorylated- to total eIF4E between the samples (*indicates total eIF4E is too low for a reliable analysis).
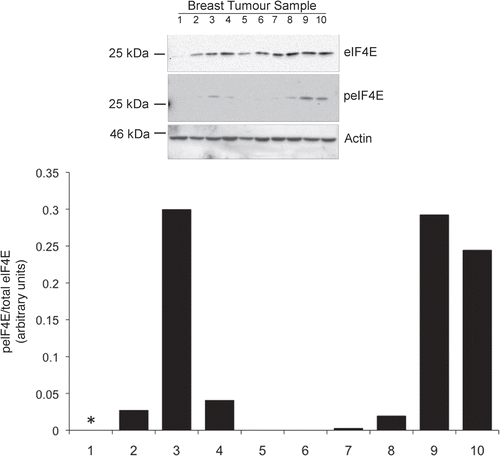
Figure 2 eIF4E expression and phosphorylation in breast cancer tissue. Cells in exponential growth phase were lysed 24 h after the application of fresh medium. Proteins were separated by SDS-PAGE and western blots probed with antibodies to MNK1, eIF4E and serine 209 phosphorylated eIF4E. Figure representative of two independent blots. Quantitation is as per .
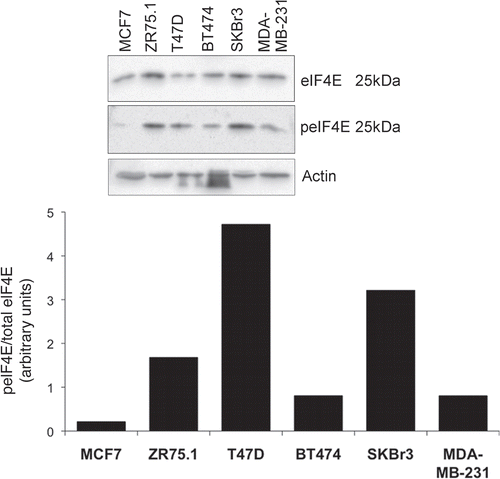
Figure 3 Dose response analysis of the inhibition of eIF4E phosphorylation by CGP57380. BT474 and SKBr3 cells were treated with the indicated concentrations of CGP57380 for 24 h prior to analysis of cell lysates by western blotting.
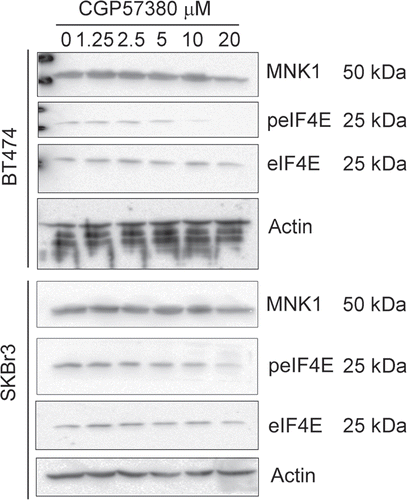
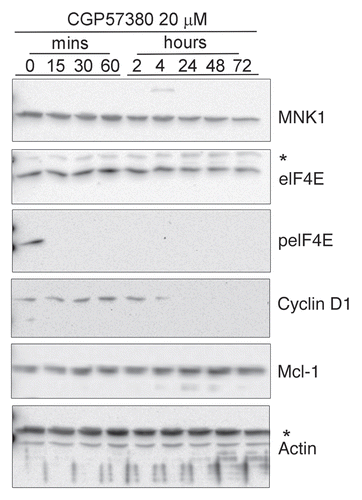
Figure 4 Time course analysis of the inhibition of eIF4E phosphorylation by CGP57380. Cells were treated with 20 µM CGP57380 for the times as indicated. Proteins were analysed by western blotting. *, cross reacting band.
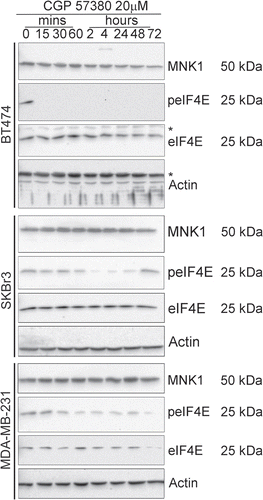
Figure 5 Effects of the MNK inhibitor, CGP57380, on colony formation by breast cancer cell lines. Cells were treated with CGP57380 20 µM or DMSO vehicle control for 24 h before being trypsinised, counted and re-plated at equal number in CGP57380 or DMSO. Colonies were fixed with methanol and stained with Giemsa 9-12 days later. Representative of three replicate plates.
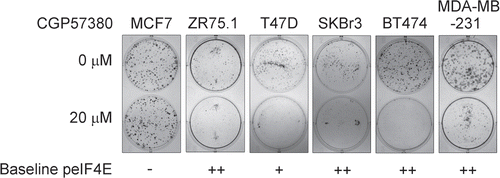
Figure 6 Effects of 72 h exposure to CGP57380 on breast cancer cell line proliferation. Cells for the experiment were plated at a density which resulted in them being in exponential growth phase at the point of assay. Data represents the results from three independent experiments for each cell line. (Bars = mean + SEM, X axis labels represent CGP57380 concentration (µM) 0 = 0.5% DMSO carrier control).
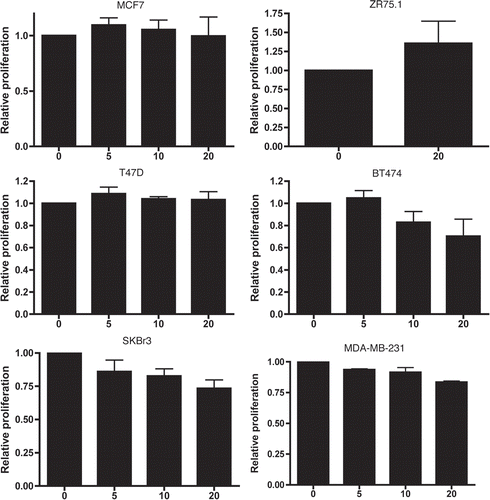
Figure 7 Live cell imaging analysis of CGP57380 treated breast cancer cells. MDA-MB-231 cells and SKBr3 cells were treated with CGP57380 20 µM or DMSO vehicle control before undergoing time lapse photomicroscopy. Graphs show the number of mitotic (black bars) and apoptotic events (white bars) per 100 initial cells during the 24 h imaging period. Results are mean of three experiments + SEM.
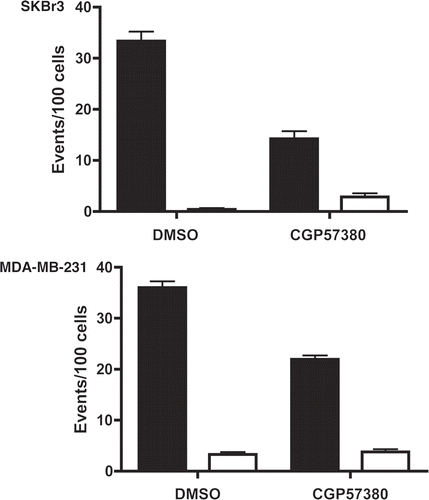
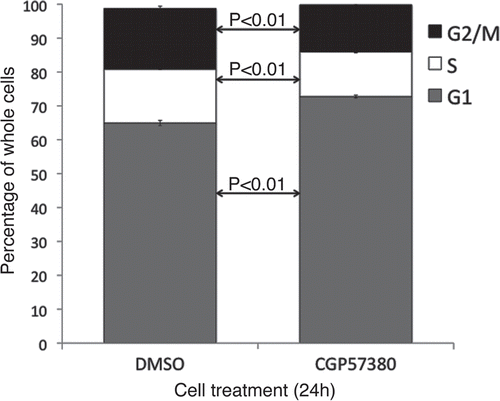
Figure 8 Cell cycle analysis of MDA-MB-231 cell line following CGP57380 treatment. MDA-MB-231 cells were treated with CGP57380 20 µM or DMSO (vehicle control); for 24 h prior to analysis of DNA content by propidium iodide staining and flow cytometry. Data shown is the means of three independently repeated experiments, each comprising three replicates. Student's t-test was used for statistical analysis.