Figures & data
Figure 1 Zebrafish hdac1hi1618 mutation impairs exocrine pancreas development by reducing epithelial proliferation, perturbing morphogenesis and inducing acetylation of nucleosomal histones H3 and H4. (A) Exocrine pancreas (green arrow) by in situ hybridization using trypsin anti-sense riboprobes in hdac1hi1618 mutant larvae and WT siblings from 48 to 120 hpf. The images represent dorsal view of the larvae with exocrine pancreas located on the right side. (B) Acinar cytodifferentiation and morphology of exocrine pancreas in hdac1hi1618 mutant larvae and WT siblings at 72 hpf by immunohistochemistry using anti-Cpa antibodies (red) and anti-cadherin antibodies (green), respectively, followed by transverse histological sectioning. The nuclei were stained with DAPI (blue). e.p., exocrine pancreas; i, intestine. Note that, in the hdac1hi1618 mutant, the morphology of the intestine is grossly normal but relatively small. (C) Proportion of exocrine pancreatic epithelia in S phase of cell cycle in hdac1hi1618 mutant larvae and WT siblings at 72 hpf by immunohistochemistry using anti-BrdU antibodies, followed by transverse histological sectioning. The number (#) of DAPI+ nuclei, the number of BrdU+ nuclei (cells in S phase) and the proportion of BrdU+ nuclei are represented as the mean ± standard deviation. *p < 0.05 indicates statistical significance. Five larvae in each group were scored. (D) Immunoblot analysis of acetylated histones H3 and H4 in hdac1hi1618 mutants and WT siblings at 72 hpf. Anti-total histones H3 and H4 antibodies and anti-actin antibodies were used as control.
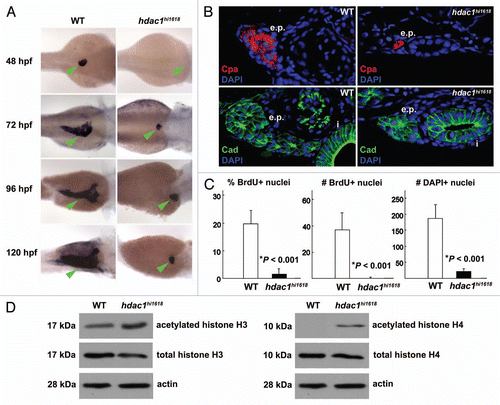
Figure 2 Anti-sense inhibition of hdac1 expression recapitulates defects of exocrine pancreas in hdac1hi1618 mutants. Exocrine pancreas of 72 hpf WT larvae injected with hdac1-MO or hdac1-5-mispair-MO analyzed by (A) in situ hybridization using trypsin anti-sense riboprobes; (B) by immunohistochemistry using anti-cadherin antibodies (green) and DAPI (blue) followed by histological sectioning. e.p., exocrine pancreas; i, intestine. Note that both exocrine pancreas and intestine are hypomorphic in the hdac1-MO-injected larvae. (C) Immunoblot analysis of acetylated histones H3 and H4 in hdac1-MO or control MO-injected WT larvae at 72 hpf. Anti-total histones H3 and H4 antibodies and anti-actin antibodies were used as controls.
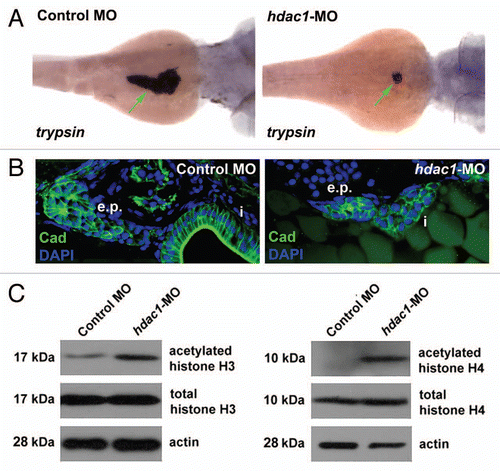
Figure 3 Expression of p21cdkna1 and hhip mRNA is upregulated in the hdac1hi1618 mutants. Semi-quantitative analysis of (A) p21cdkn1a and (B) hhip mRNA in hdac1hi1618 mutants and WT siblings at 48 hpf by real-time PCR using gapdh mRNA as internal control. Each column represents the mean relative value of mRNA level using RNA extracted from three independent groups of hdac1hi1618 mutant and WT larvae, with triplicates per sample for real-time PCR. Bars represent mean ± SD. *p < 0.05 indicates statistical significance.
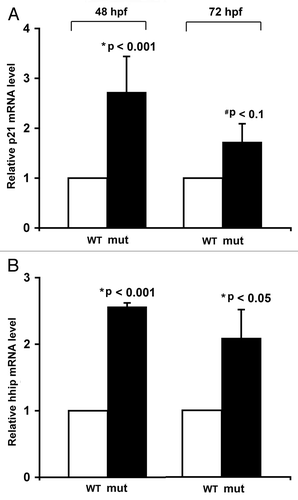
Figure 4 Expression of HDAC1 is upregulated in human pancreatic adenocarcinoma tissues and cells. (A and B) Immunohistochemical analysis of HDAC1 protein in normal pancreatic tissue and pancreatic adenocarcinoma obtained from different regions of the pancreas from the same patient. These images are representative of the normal-tumor pairs of pancreatic tissues from five patients. Red arrows point at the nuclei of the ductal cells. Pre-immune serum was used as control for specificity of anti-HDAC1 antibodies (data not shown). (C) Quantitative analysis of HDAC1 mRNA in human pancreatic adenocarcinoma cell lines by real-time PCR. Each value represents the relative HDAC1 mRNA level in the cell line as compared with that in H6c7. Each column represents the mean and the bars represent SD, as determined from three independent experiments. *Indicates statistically significant difference relative to H6c7.
Figure 5 RNA interference-induced silencing of HDAC1 reduces proliferation of pancreatic adenocarcinoma cells by impairing cell cycle progression. Panc 02.03 cells treated with anti-HDAC1 siRNA, non-specific control siRNA or no siRNA were analyzed. (A) Bright field images with phase contrast at 24 h post-transfection. Multi-nucleated cells are indicated by arrows and cytoplasmic vacuoles by arrowheads. (B) Proliferation assays by MTS colorimetric absorbance and by counting viable cells. Columns and bars represent the mean and SD, respectively. (C) Cell cycle by FACS analysis of DNA content. The proportion of cells in each phase of the cell cycle is indicated. Non-specific sub-G0/G1 events were gated out of analyses, allowing for more accurate analysis of cell cycle distribution curves. These experiments were repeated with essentially the same results.
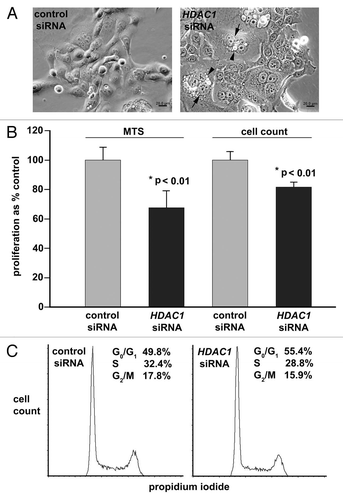
Figure 6 Targeted knock down of HDAC1 induces acetylation of nucleosomal histones and upregulates expression of CDK inhibitors and SHH signaling components. Panc 02.03 cells were transfected with anti-HDAC1 siRNA or non-targeting control siRNA. Total protein and RNA were extracted and analyzed at 48 h post-transfection. (A) Immunoblot analysis of acetylated histones H3 and H4 and total histones H3 and H4. The immunoblots are representative of two independent experiments with essentially the same results. (B) Semi-quantitative analysis of p21CDKN1A, p27CDKN1B, HHIP and SMO mRNA by real-time PCR. Columns and bars represent mean and SD, respectively for mRNA of each gene in the cells transfected with anti-HDAC1 siRNA as % of those in the control siRNA-transfected cells. *p < 0.05 indicates statistical significance.
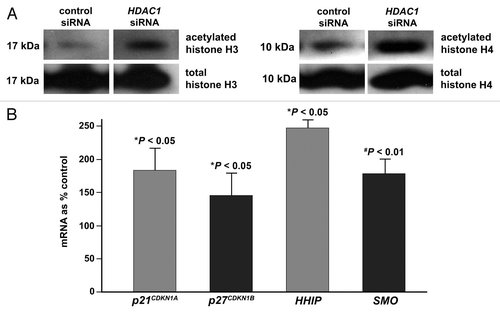
Table 1 Selected transcriptional profile of hdac1hi1618 mutants as compared to WT siblings using RNA extracted at 48 hpf