Figures & data
Figure 1. Resveratrol-induced apoptosis is inhibited by U0126. (A) Cell lysate from LNCaP and C4–2 cells, treated with the indicated doses of resveratrol for 12 h in serum-free conditions, were blotted with the indicated antibodies. The two cell lines exhibited differential cleavage of PARP. (B) Nucleosome ELISA data confirm differential apoptosis in LNCaP and C4–2 cells at 2 μM resveratrol after 12 h exposure. (C) Cell lysates from C4–2 cells treated with the indicated MEK inhibitors, at the indicated doses of resveratrol, were subjected to protein gel blot. PARP cleavage is selectively inhibited by 10 μM U0126. (D) U0126 inhibition of apoptosis by resveratrol is confirmed by nucleosome ELISA at 12 h. (E) Cell lysates from C4–2 cells treated with the indicated MEK inhibitors after EGF stimulation in serum-free conditions.
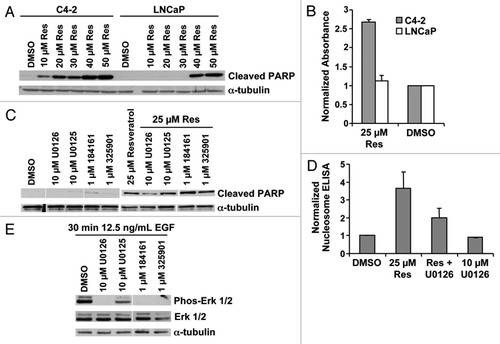
Figure 2. U0126 induces mitochondrial fluorescence. (A) Fluorescence imaging of LNCaP cells treated with U0126 for 12 h and stained with the mitochondrial dye Mitotracker CMXRos. Note the FITC perinuclear signal that colocalizes with the mitochondrial stain. (B) Fluorescence localization in the mitochondria is confirmed by cell fractionation and fractionation purity is assayed by protein gel blotting of compartment specific markers: α-tubulin for cytoplasm (cyto), MnSOD for mitochondria (mito), and lamin A/C for nucleus (nuc). (C) Multiple MEK inhibitors did not produce a fluorescent signal when incubated in C4–2 cells for 12 h. (D) U0126 produced fluorescence in multiple mammalian cells after 12 h incubation and (E) in BY4741 yeast.
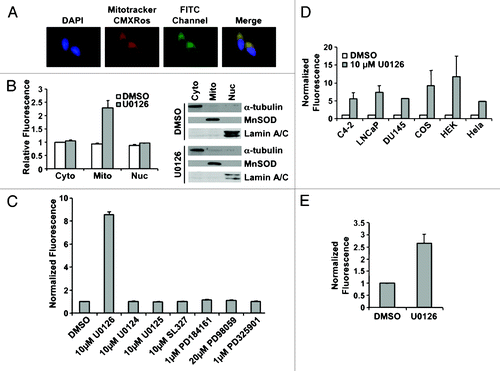
Figure 4. Biological consequences of mitochondrial inhibition by U0126. (A) Cell morphology of C4–2 cells 48 h after drug treatment. U0126, U0124 were dosed at 10uM and PD325901(PD32) was dosed at 1 μM. (B) Crystal violet staining of C4–2 and T24 cells at 48 h after treatment with DMSO (D) or U0126 (U) and varying concentrations of sodium oxamate (Ox). (C) Viable cell counting with trypan blue of C4–2 cells 48 h after treatment with the indicated drugs. (D) Lactate dehydrogenase (LDH) enzyme activity from cell lysates incubated with varying concentrations of sodium oxamate to verify LDH inhibition.
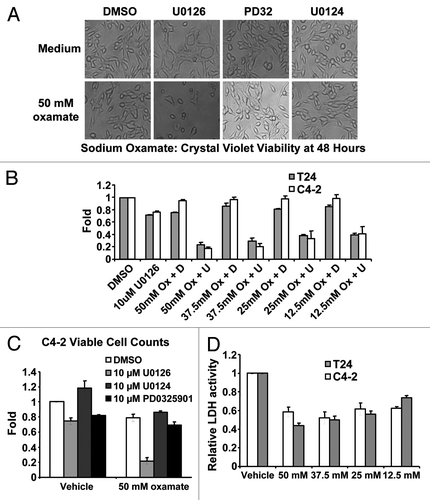
Figure 3. U0126 induces a metabolic shift in prostate cancer cells. (A) Lactic acid levels in the medium of LNCaP and C4–2 cells were assayed 24 h after treatment with U0126. DMSO was used as a vehicle. (B) Mitochondrial potential was assayed at 12 h after dosing various MEK inhibitors in C4–2 cells. (C) Cellular ATP levels were measured 9 h after treatment with U0126. Prior to assay, cells were preincubated with 10mM 2-deoxygluocse for 30 min to inhibit glycolytic ATP production, but not mitochondrial production. (D) Cyanide levels detected in cell lysates exposed to different concentrations of U0126 for 24 h. **p < 0.05, two-tailed Student’s t-test.
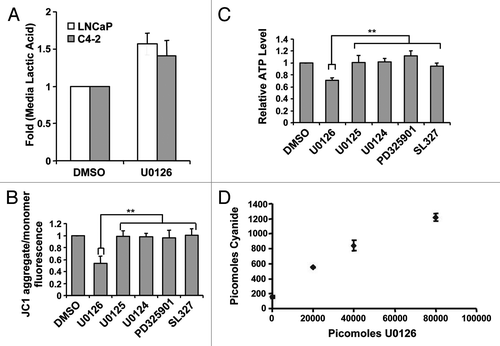
Figure 5. Role of glutamine metabolism in resveratrol induced apoptosis. (A) Transaminase inhibitors AOA and cycloserine were used in serum-free conditions. Twelve hours later resveratrol was added and cell death was assayed at 24 h by cleaved PARP or (B) nucleosome ELISA. **p < 0.05, paired two tailed t-test. (C) Cell death induced by resveratrol was assayed at varying glutamine concentrations. (D) Cell viability by crystal violet staining 60 h after resveratrol treatment at varying glutamine concentrations. **p < 0.05, paired one tailed t-test (E) The non-metabolizable glutamine analog, 6-diazo-5-oxo-L-norleucine (DON), did not restore resveratrol-induced death in glutamine-free medium.
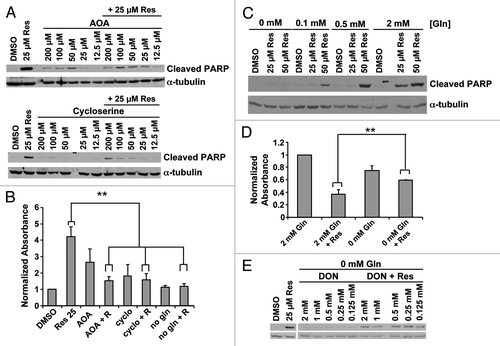
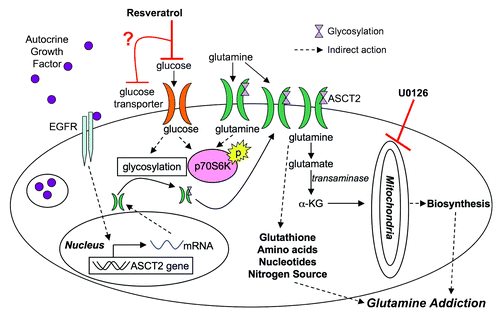
Figure 6. Model for resveratrol action in cancer metabolism. Based on the results presented, resveratrol may inhibit glucose uptake leading to a cascade of downstream effectors. Glutamine metabolism may be adversely targeted by downregulation of ASCT2 from the surface of the cell, depleting glutamine levels in the cell, which ultimately induce apoptosis in cancer cells (α-KG, α ketoglutarate).