Figures & data
Figure 1 Cytotoxicity was induced by Lip-C6, Lip-PDMP and gemcitabine in highly drug-resistant PANC-1 pancreatic cancer cells. Cellular viability of PANC-1 cells was determined at 24, 48 and 72 h in a dose response utilizing: (A) Lip-C6, (B) Lip-PDMP and (C) gemcitabine (Gem). (D) IC50 values for individual treatments at 48 h were calculated. All data points are representative of n = 8 experimental conditions.
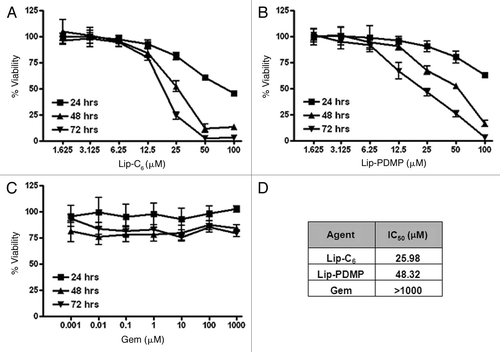
Figure 2 Lip-C6, Lip-PDMP and gemcitabine cooperatively induce apoptosis of PANC-1 cells. Apoptosis of PANC-1 cells was detected by TUNEL assay following 24 h treatments with: (A) saline control, (B) Lip-C6 (5 µM C6-ceramide), (C) 20 µM gemcitabine (Gem), (D) Lip-C6 (5 µM C6-ceramide) + 20 µM Gem, (E) Lip-Ghost (empty nanoliposome), (F) Lip-PDMP (5 µM PDMP), (G) Lip-C6/PDMP (5 µM C6-ceramide and 5 µM PDMP), and (H) Lip-C6/PDMP (5 µM C6-ceramide and 5 µM PDMP) + 20 µM Gem. (I) Apoptotic cells were quantified as a percent of the total cell number. One-way ANOVA: *p < 0.001 compared with control, Lip-Ghost, Lip-C6, Gem and Lip-PDMP, #p < 0.05 compared with Lip-C6 + Gem and Lip-C6/Lip-PDMP, n = 5.
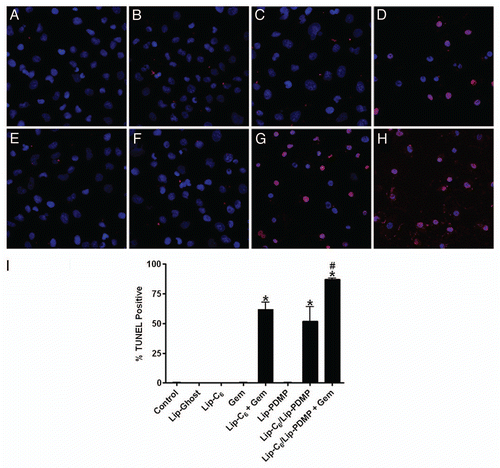
Figure 3 The metabolic fate of Lip-C6 is altered by Lip-PDMP alone or in combination with gemcitabine. PANC-1 cells were treated for 24 h with 12.5 µM Lip-C6, 24 µM Lip-PDMP, 40 µM gemcitabine (Gem) or various combinations. Cells were harvested and lipids were extracted and analyzed using LC-MS/MS/MS. Abundance relative to total cellular protein was determined for: (A) C6-ceramide, (B) C6-cerebroside, (C) C6-sphingosmyelin, (D) total natural (endogenous) ceramide, (E) sphingosine and (F) sphingosine-1-phosphate. One-way ANOVA: *p < 0.05 compared with control and Lip-Ghost, #p < 0.05 compared with Lip-Ghost only, $p < 0.05 compared with Lip-C6 (Lip-C6-containing combinations only), %p < 0.05 comparing triple combination with Lip-C6 + Gem only, &p < 0.05 comparing triple combination with Lip-C6 + Lip-PDMP only, @p <0.05 comparing triple combination with Lip-C6 + Gem and Lip-C6 + Lip-PDMP, n = 4.
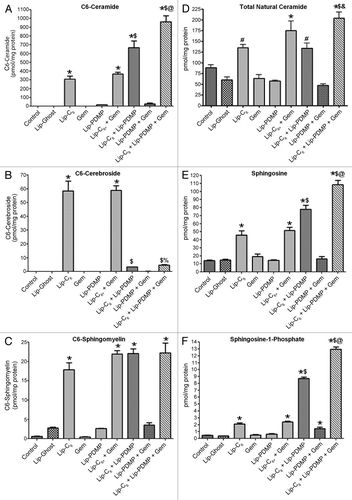
Figure 4 Pharmacological inhibition of Akt or Erk in PANC-1 cells replicated the effect of Lip-C6 on these signaling pathways. PANC-1 cells were exposed to the Akt inhibitor SH-6 or the MEK inhibitor U0126 (a kinase upstream of Erk). (A) Phosphorylation of Akt was blocked by 48 h treatment with SH-6 (9.5 µM), and phosphorylation of Erk was blocked by 48 h treatment with U0126 (17.5 µM). (B) Cellular viability was determined at 48 h in a dose response utilizing SH-6. (C) Cellular viability was determined at 48 h in a dose response utilizing U0126. (D) The effects of SH-6 (4.25 µM) on cellular viability were compared with Lip-C6 (25 µM) treatment or were evaluated in combination. (E) The effects of U0126 (17.5 µM) on cellular viability were compared with Lip-C6 (25 µM) treatment or were evaluated in combination. One-way ANOVA: *p < 0.001 compared with Lip-Ghost + DMSO or Lip-Ghost + SH-6, **p < 0.001 compared with Lip-C6 + DMSO, n = 8.
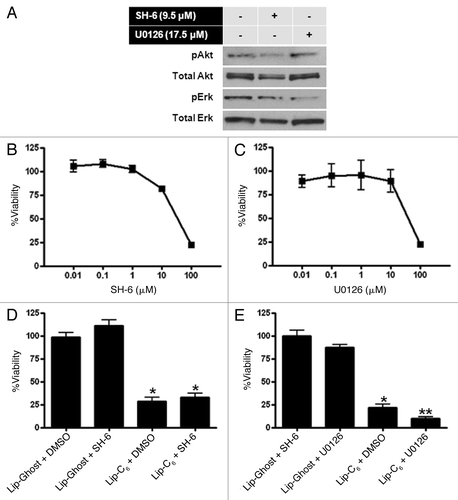
Figure 5 Lip-C6, but not gemcitibine, inhibits Akt and Erk signaling pathways in PANC-1 cells. PANC-1 cells were maintained in media containing 2.5% FBS to reduce the background level of phosphorylation. Cells were treated with control (media only), Lip-Ghost (total lipid weight-matched), Lip-C6 (35 µM C6-ceramide), 20 µM gemcitabine (Gem), or a combination of Lip-C6 and Gem, for 24 h. Cells were harvested, lysed and total proteins were subjected to protein gel blotting. (A) Phosphorylated-Erk (pErk) and (B) phosphorylated-Akt (pAkt), were detected using monoclonal antibodies against pErk and pAkt, respectively. Total Erk and total Akt, protein levels were quantified using anti-Erk and anti-Akt, antibodies, respectively one-way ANOVA: #p < 0.01 compared with control or Gem, *p < 0.05 compared with control, Lip-Ghost or Gem, n = 3).
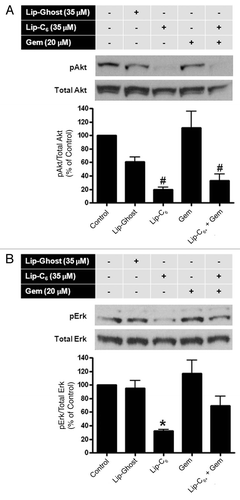
Figure 6 The in vivo antitumor efficacy of Lip-C6 is augmented by gemcitabine or Lip-PDMP. Bilateral subcutaneous PANC-1 tumors were established on the flanks of athymic nude mice. (A) Lip-Ghost (equivalent total lipid dosage), Lip-C6 (9 mg/kg C6-ceramide), 50 mg/kg gemcitabine (Gem) or a combination of Lip-C6 and Gem, were routinely administered via tail vein injection. Two-way ANOVA: *p < 0.05, Lip-Ghost compared with all other treatments (large box over days 36 to 54), #p < 0.05, Lip-Ghost compared with Lip-C6 + Gem (small boxes over day 54), n ≥ 4. (B) Lip-Ghost (equivalent total lipid dosage), Lip-C6 (18 mg/kg C6-ceramide) or Lip-C6/PDMP (18 mg/kg C6-ceramide and 23 mg/kg PDMP), were routinely administered via tail vein injection. Two-way ANOVA: **p < 0.05, Lip-Ghost compared with Lip-C6/PDMP (small boxes over days 60–63), n ≥ 4.
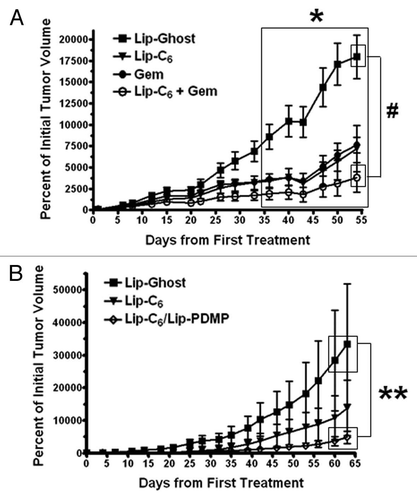
Table 1 Nanoliposome formulations
Table 2 Synergy of combinatorial therapies
Table 3 The conversion of Lip-C6-delivered C6-ceramide to natural ceramide species is altered by Lip-PDMP alone or in combination with gemcitabine