Figures & data
Figure 1 Efficient construction of 5FdU containing heteroduplex plasmid. (A) Flowchart of for preparation of the 5FdU containing heteroduplex plasmid. (B) Electrophoretic analysis of various DNA preparations. Lane 1; parental pGEM7Zf(+) dsDNA plasmid isolated by an alkaline lysis method. Almost of all the plasmid was in supercoiled form. Lane 2; single-stranded DNA plasmid generated by conventional M13KO7 method (17). Lane 3; Single-stranded DNA isolated from JM109/pGEM7Zf(+)/M13KO7 complex. Lane 4; 5FdU-containing heteroduplex plasmid constructed by ssDNA plasmid generated by conventional M13KO7 method. Lane 5; 5FdU-containing heteroduplex plasmid constructed by ssDNA plasmid isolated from JM109/pGEM7Zf(+)/M13KO7 complex. Lane 6; linear dsDNA marker (1 kb Ladder N3232S, New England Biolabs). We generated less contaminated ssDNA by the bacteria/phagemid/helper phage complex method than by the conventional M13KO7 infection method (17) (lane 2 and 3), which also reduced the amount of contamination after the oligomer extension reaction (lane 4 and 5). (C) Sequence at the heteroduplex site. PCR was performed using the constructed plasmid as a template. After purifying the PCR product, direct sequencing was performed to confirm the heteroduplex site. KM, kanamycin; Amp, ampicillin; Nicked, nicked dsDNA plasmid; Supercoiled, supercoiled plasmid.
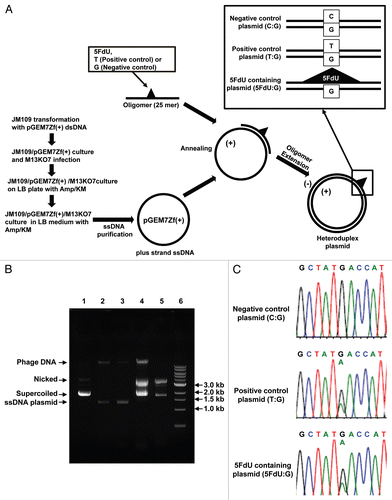
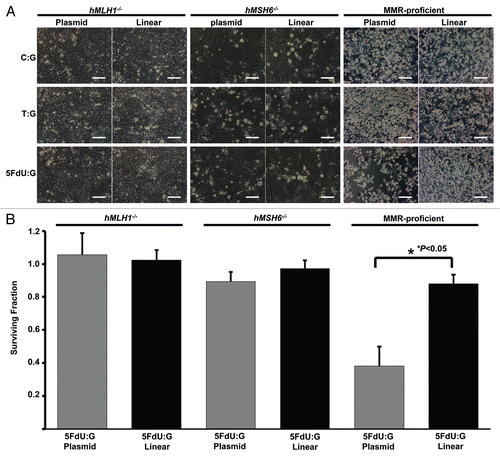
Figure 2 5FdU-containing heteroduplex plasmid leads to cellular morphological changes in MMR-proficient cells. Twelve hours after transfection, hMLH1-/- (A), hMSH6-/- (B) and MMR-proficient (C) cells were seeded at a density of 1.5 × 105 cells per well onto 6-well plates and observed by microscope. By 24 h after seeding, the 5FdU plasmid induced morphological changes consistent with cell death in the MMR-proficient cells (C), while no morphological changes were observed in the hMLH1-/- (A) and hMSH6-/- (B) cells. Neither MMR-proficient cells nor MMR-deficient cells demonstrated morphological changes by heteroduplex (T:G) and negative (C:G) control plasmids. Scale bars; 10 µm. 5FdU:G; 5FdU containing heteroduplex plasmid, T:G; positive control heteroduplex plasmid, C:G; unaltered plasmid (negative control).
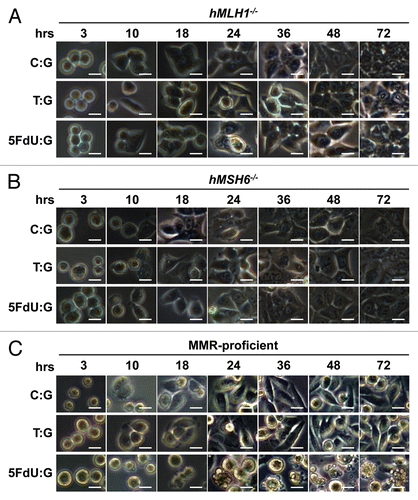
Figure 3 5FdU containing heteroduplex plasmid induces cytotoxicity in MMR-proficient cells. (A) MTS assay. Cells (5,000 cells/well) of each population were seeded in 100 µl of culture medium in the wells of three 96-well plastic plates and plates were incubated in a 5% CO2/95% air incubator for 24 , 48 and 72 h. After incubation, 10 µl of MTS reagent solution was added to each well, and the plate was incubated for an additional 4 h in the 5% CO2/95% air incubator. Cell viability was measured by scanning with a microplate reader at 490 nm. This experiment was performed three times. (B–D) Clonogenic assay. Twelve hours after transfection, hMLH1-/- (B), hMSH6-/- (C) and MMR-proficient (D) cells were seeded at a density of 5 × 102 cells/100 mm dish and colonies were stained with Giemsa and counted 14 days after seeding. Survival fraction was shown as a percentage of negative controls. This experiment was performed at least three times.
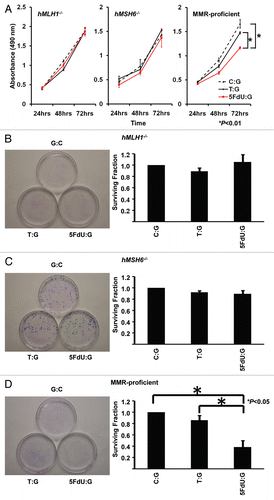
Figure 4 5FdU-containing heteroduplex plasmid is more useful for measurement of cytotoxicity than linear dsDNA. (A) Cell density. Twelve h after transfection, cells were seeded at a density of 1.5 × 105 cells per well into 6-well plates. 72 h after seeding, the cell density was observed by microscopy. 5FdU-plasmid transfected MMR-proficient cells showed lower cell density than 5FdU linear dsDNA transfected cells. (B) Clonogenic assay. Twelve hours after transfection, cells were seeded at a density of 5 × 102 cells/100 mm dish and colonies were stained with Giemsa and counted 14 days after seeding. Survival fraction was shown as a percentage of negative controls. This experiment was performed at least three times. Scale bars; 100 µm.