Figures & data
Figure 1. Values are the mean percentage (%) (± Standard Deviation) of dead/dying A431 cells measured 24 h after irradiation of three independent experiments. The cells were incubated for 1 h with Nimotuzumab (gray circles) and Cetuximab (black squares) mAbs and exposed to ionizing radiation (137Cs, 4 Gy). *ANOVA and Student t-test on slope, p < 0.05. In spite of the cytotoxic response was 1.7 times greater for cetuximab than for nimotuzumab, there was not a statistical difference between then (p > 0.05).
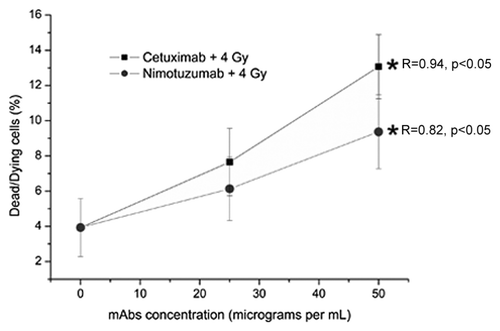
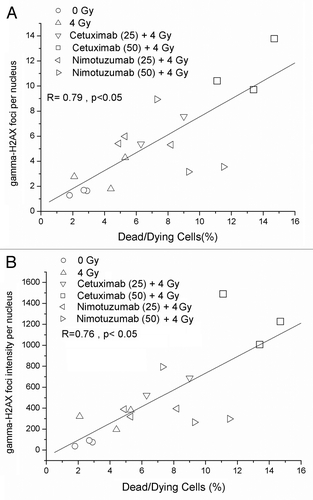
Figure 2. Detection of residual DNA damage in A431 cells measured 24 h after irradiation (137Cs, 4 Gy). The cells were incubated for 1 h with Nimotuzumab (gray circles) and Cetuximab (black squares) and exposed to ionizing radiation (137Cs, 4 Gy). (A) γ-H2AX foci intensity per nucleus. (B) γ-H2AX foci number per nucleus. Values are means ± standard deviation of three independent experiments. *ANOVA and Student t-test on slope, p < 0.05.
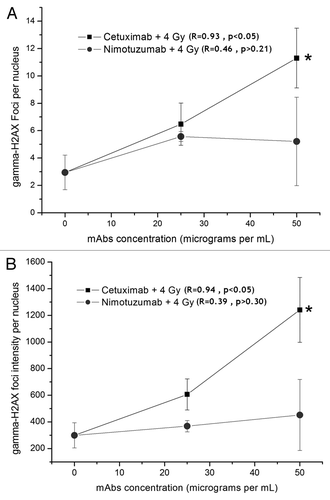
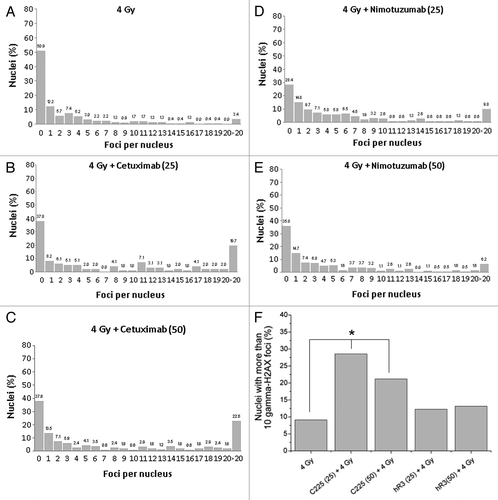
Figure 3. (A) Distribution of number of foci per nuclei after irradiation (137Cs, 4 Gy). (B) Nimotuzumab (25 µg/mL) pretreatment plus 4 Gy. (C) Nimotuzumab (50 µg/mL) pretreatment plus 4 Gy. (D) Cetuximab (25 µg/mL) pretreatment plus 4 Gy. (E) Cetuximab (50 µg/mL) pretreatment plus 4 Gy. (F) Percentage of nuclei containing more than 10 foci. *Chi-square trend test, p < 0.05.