Figures & data
Figure 1 Elevated EP4 receptor expression in human colon cancer specimens. Immunohistochemistry (IHC) staining of EP4 receptor protein was performed in human colon tissue specimens. (A) The staining of normal colonic mucosa shows low EP4 receptor protein expression on the apical membrane. (B) Elevated EP4 staining was observed in adenocarcinomas lesions. (C) Differences between IHC scores for normal, adenomatous, and cancerous lesions are shown as a box and whisker plot. The distribution of pair-wise comparisons of differences in EP4 staining scores shows a significant difference in EP4 receptor staining in malignant colonic lesions compared with normal and adenoma lesions (*p < 0.0001). Each dot represents IHC score (intensity of stain X percentage of cells stained) generated after staining formalin-fixed paraffin embedded tissues for EP4 antibody by IHC.
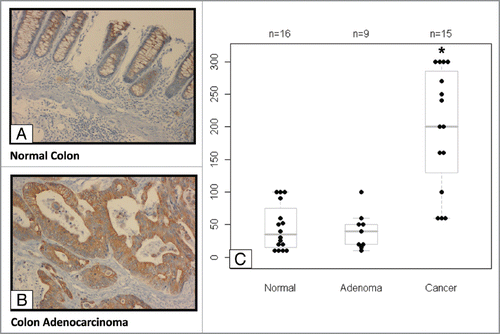
Figure 2 Highly conserved miR-101 seed sequence within the PTGER4 mRNA. (A) Computational prediction of miR-101 seed region on PGTER4 mRNA. The miR-101 target sequences are located on nucleotide 18–24 in 3′-UTR of PTGER4. The target region is denoted with bold letters. (B) The miR-101 seed sequence is highly conserved across five species.
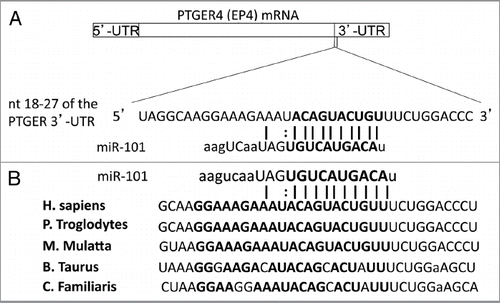
Figure 3 Inverse relationship between EP4 receptor protein and miR-101 expression levels in colon cancer tissues. (A) Western analysis characterizing EP4 receptor protein levels in different colon cancer cells lines. (B) Characterization of miR-101 expression levels in various colon cancer cell lines using qRT-PCR. (C) Western analysis of EP4 receptor protein isolated from paired normal (N) and colon tumor (T) tissue specimens. Tumor colon specimens display higher EP4 receptor protein levels. (D) qRT-PCR analysis showing relative levels of miR-101 in paired normal colon and cancer specimens. The miR-101 expression levels of tumors (T) were normalized against their corresponding levels in normal (N) colon tissues. Note, loss of miR-101 in the tumor specimens is associated with increased EP4 protein expression. **p < 0.01.
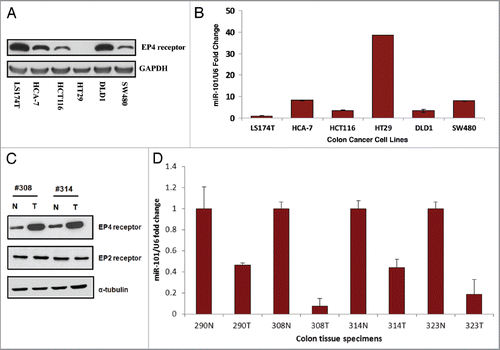
Figure 4 Analysis of miR-101 expression in human colon tissue by in situ hybridization. (A) (a) Paraffin embedded, formalin-fixed non-malignant colon epithelial tissue stained with H&E. (b) Non-malignant colon tissue hybridized with LNA scrambled probe (c) Non-malignant human colon tissue hybridized with LNA miR-101 probe. Note miR-101 expression was observed in normal colon epithelial cells and surrounding tissue. (d) Colon adenocarcinoma tissue stained with H&E. (e) Colon cancer tissue hybridized with LNA scrambled probe. (f) Colon cancer tissue hybridized with LNA miR-101 probe. Malignant adenocarcinoma cells show considerably low miR-101 expression levels or none. Images captured at 20x magnification. (B) (a) DAPI-labeled nuclei. (b) miR-101 expression detected with HRP-DIG-labeled miR-101 probe reacted with tyramide conjugated fluorescein. (c) EP4 expression revealed with HRP-EP4 antibody reacted with tyramide-rhodamine. (d) Co-detection of EP4 and miR-101. Two cell populations are visible in the malignant epithelium, cells co-expressing low miR-101 levels permitting significant EP4 expression (white arrow) and cells that had shut down miR-101 expression allowing high levels of EP4 expression (yellow arrowheads). A third group of cells expressing high miR-101 levels and no EP4 expression is identifiable surrounding the malignant epithelium (white arrowheads). De-convoluted images captured at 40x (water).
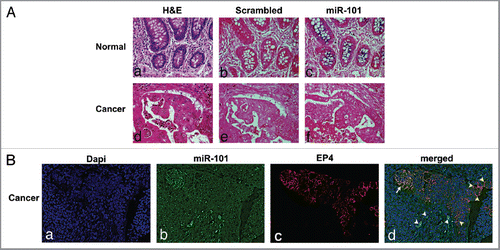
Figure 5 The EP4 receptor is a direct target of miR-101. (A) Predicted miR-101 binding sequence in the human PTGER4 3′-UTR, encoding EP4 receptor. The PTGER4 wildtype and mutant constructs were cloned into the pGL3 plasmid and downstream of the Luciferase gene at the Xba1 restriction site. The location of the wild-type constructs in the plasmid is denoted with yellow and mutant with red. Ninety percent of the seed sequence is mutated in the mutant construct. (B) The LS174T colon cells were co-transfected with pGL3carrying the constructs (wild-type or mutant EP4-3′-UTR and either a miR-101 expressing plasmid or empty vector, and Renilla. Co-transfection of the wildtype construct and miR-101 suppressed EP4 Luciferase expression. The relative luciferase activities were measured and normalized against their control. (C) Western analysis of endogenous EP4 receptor protein expression levels after ectopic miR-101 expression in LS174T cells. The levels of endogenous EP4 receptor protein decreases in a time-dependent manner
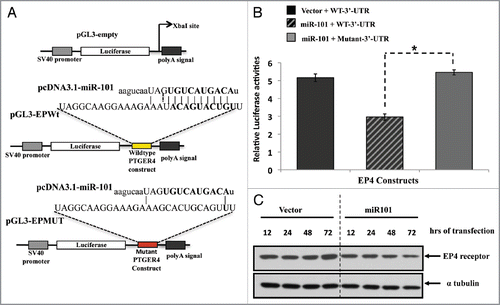
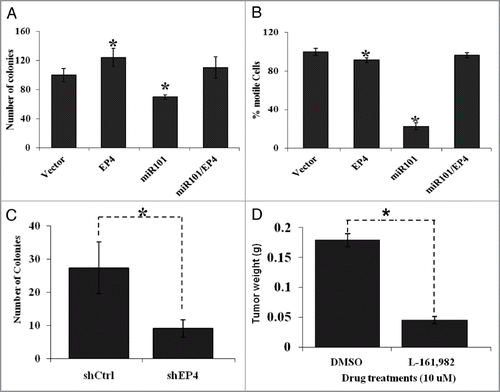
Figure 6 The biological effects of miR-101 and EP4 receptor inhibition in vitro and in vivo. (A) Ectopic expression of miR-101 in LS174T cells suppressed EP4 expression colon tumor cell motility, whereas co-expression of EP4 could overcome miR-10 suppression of motility. (B) Enforced expression of miR-101 attenuated LS174T cell motility, whereas co-expression of EP4 rescued cells from the inhibitory effects of miR-101. (C) RNAi-based silencing of EP4 receptor expression using shRNA (shEP4) reduced colony formation relative to cell containing vector scrambled sequence (shCtrl). (D) Administration of the EP4 receptor antagonist LS161982 inhibited the growth of colon cancer cells (LS174T) grown in the chorioallantoic membrane (CAM) assay. **p < 0.01.